Author: Kevin Spear
One of DNA’s elegant features is that the nucleotide bases are linked together with hydrogen bonds. Unlike their name implies, hydrogen bonds aren’t really chemical bonds at all. Instead, they are very strong attractions that exist between a hydrogen atom on one molecule and another atom (typically oxygen or nitrogen) on another molecule. Because hydrogen bonds are not true chemical bonds, they are not nearly as strong as chemical bonds. As a result, they can be “broken” with only a small amount of energy.
It turns out that this is the perfect design, because in order for DNA to code for proteins, the double helix must “open up” to expose the nucleotide bases. To do this, the link between the nucleotide bases must be broken. If the nucleotide bases were held together with chemical bonds, it would take a lot of energy to break the link, and that energy could easily damage the other bonds in DNA. Since the nucleotide bases are linked with hydrogen bonds, however, it takes only a small amount of energy to break the link. As a result, DNA can “open up” very easily, and the rest of the molecule is not harmed when that happens.
Watson and Crick determined all this, including exactly where the hydrogen bonds formed. Not surprisingly, the way in which the links form is called Watson-Crick pairing. Well, it turns out that there is another way the nucleotide bases can pair up, and a recent study shows that this is yet another amazing design feature of DNA.
Ten years after Watson and Crick proposed their model, Karst Hoogsteen was trying to confirm how the nucleotide bases linked together by studying adenine and thymine in solution. However, he found that they linked together in a different way. The way they linked together in his experiment is called, not surprisingly, Hoogsteen pairing.1 The difference between the two can be seen by the following illustration, which shows the hydrogen bonds with dotted lines:
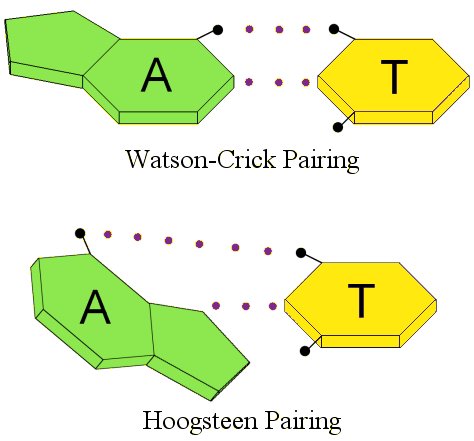
Notice the difference. The nucleotide base adenine (A) is “flipped” in Hoogsteen pairing as compared to Watson-Crick pairing. While this doesn’t look very different, remember that the nucleotide bases are connected to the DNA backbone. One nucleotide base is connected to one helix, and the other is connected to the other helix. If I flip one of those nucleotide bases, that’s going to actually change the shape of that portion of the DNA!
Well, it turns out that the majority of nucleotide bases in DNA link together with Watson-Crick pairing. However, there are a few that link together with Hoogsteen pairing. It has been thought for quite some time that Hoogsteen pairing only occurs in DNA when it is either damaged or bound to some other molecule (like a protein or a drug). A recent study, however, shows that this is not the case. In fact, Hoogsteen pairing seems to be a part of DNA’s design.
Evgenia Nikolova and colleagues used some advanced nuclear magnetic resonance (NMR) techniques to study Hoogsteen pairing, and they found that it happens routinely in DNA. Although it happens only about 1% of the time, they show that it is actually functional. They end up referring to such pairing as an “excited state” for DNA.2 Thus, far from being an indication of damage or external binding, it seems to be a means by which DNA increases it complexity so as to store more information. The authors state that their observations imply
that the DNA double helix intrinsically codes for excited state Hoogsteen base pairs as a means of expanding its structural complexity beyond that which can be achieved based on Watson–Crick base-pairing.
In other words, Hoogsteen pairing gives DNA yet another way to store information! If this study is confirmed, then, there is yet another layer of complexity to DNA, which up until now has been completely unknown.
The more I study God’s handiwork, the more I stand in awe of Him!
REFERENCES
1. Albert L. Lehninger, David Lee Nelson, and Michael M. Cox, Principles of Biochemistry, Fourth Edition, (W. H. Freeman, 1998), p. 286.
Return to Text
2. Evgenia N. Nikolova, et al., “Transient Hoogsteen base pairs in canonical duplex DNA,” Nature, doi:10.1038/nature09775, 2011
Return to Text

Have you seen the several ways that DNA uses to “spell check” itself during replication? I have just been reading about it and it is extraordinary! And this information comes from researching “simple” organisms like bacteria.
DNA polymerase is amazing, isn’t it? There is also a “spell check” function during translation to ensure that the gene is faithfully converted to the proper protein. It amazing the number of ways a cell can check to make sure it’s doing what it’s supposed to be doing.
Dr. Wile,
Thanks for the article. DNA is amazing! I was just curious if you knew what the information storage of DNA is vs. the Binary/Digital system we use. How much more information can DNA store while occupying the same space? Also, is there some way we could make a paradigm shift in information technologies by mimicking this form of storage? Also, this brings up another question I had: how does the brain store memory? Does it use a similar “code” as DNA or does it use some other mechanism in order to store the massive information it does?
Good questions, Enoch. DNA stores information in the form of nucleotide bases. There are (on average) 10 nucleotide bases in a single winding of DNA, and that winding occupies about 1.068×10^-19 cubic millimeters (the “^” means “raised to”). So the “information density” of DNA is about 10/1.068×10-19 = 9.4 × 10^17 nucleotide bases per cubic millimeter. The genetic code is based on 3 nucleotide bases identifying an amino acid or action, so assuming that 3 nucleotide bases is like a bit, that means you have about 3.1 x 10^17 bits per cubic millimeter.
While the number keeps increasing, the information density of most modern hard drives is about 5 x 10^6 bits per cubic millimeter. Thus, DNA stores about 3.1 x 10^17/5 x 10^6 = 62,000,000,000 times as much data per cubic millimeter as a hard drive!
There are many people trying to produce computers based on living organisms and even DNA. There are bacterial computers as well as DNA computers, but they are in their infancy and have a lot of technical hurdles to go through before they become practical.
The precise way in which the brain stores memories is not understood, but it is nothing like how DNA stores information. The “workhorse” cells of your brain are called neurons, and we know that the interconnectedness of your neurons grows over time. Thus, it is thought that the interconnectedness of the neurons facilitates memory storage. It was once thought that adults do not make new neurons in their brains, but we now know that is not true. The new neurons are small in number compared to the total number of neurons in the brain, however, so they can’t be the sole means by which memories are stored. An interesting review article from Science News a few weeks back said that current research indicates those new neurons help your brain distinguish between memories that are distinct but similar.