In a previous post, I discussed alternative splicing, an amazing aspect of our DNA that allows it to store information in a compact, elegant way. In brief, a gene is actually a recipe that the cell uses to make a particular protein. Since most of a cell’s DNA is in the nucleus, the “recipe” stored in that gene must leave the cell’s nucleus in order to be turned into a protein. To do that, the “recipe” is copied by a molecule called messenger RNA (mRNA). The mRNA then takes the copied “recipe” out of the nucleus to the ribosome, which is where proteins are made.
In eukaryotic cells (the kinds of cells found in plants and animals), however, something very interesting happens before the mRNA leaves the nucleus. Some parts of the mRNA are cut away, and the remaining parts are then stitched back together. The parts of the mRNA that are cut away never leave the nucleus, so they are called introns (they stay IN the nucleus). The remaining parts that are stitched together are called exons (they EXit the nucleus). For a while, geneticists didn’t know the purpose of introns, so in typical evolutionary fashion, many decided that they had no purpose, and introns were lumped into the category of “junk DNA.” Of course, as we have learned more about genetics, we have learned that the evolution-inspired idea of “junk DNA” is, itself, junk, although some evolutionists still cling to it.
Nowadays, of course, we know the reason that introns exist. It is part of the design of the Creator, allowing DNA to store information in an incredibly efficient way. Each exon represents a “module” of useful information. If the cell stitches the exons together in one way, it makes one protein. If it stitches the exons together in another way, it makes a different protein. As a result, a single gene can actually produce many different proteins. The introns not only serve as a means by which the cell can identify the exons, they also regulate the amount of the various proteins that are being made.
This process of alternative splicing is illustrated in the figure below:
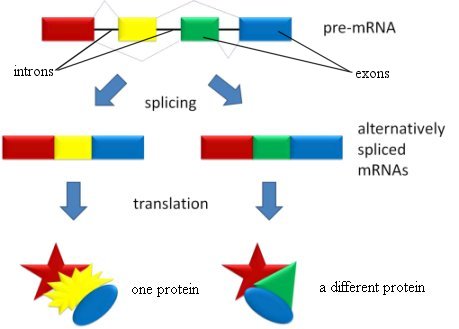
In the previous post about alternative splicing, I discussed how recent evidence suggests that up to 95% of the genes in the human genome participate in this process. However, I did not address how many different proteins a single gene can produce using alternative splicing. In some cases, the answer is truly astounding.
Over the past couple of years, I have been helping the head of the biology department at a Christian university as he works with three other creation scientists on a university-level general biology textbook. The last time we met, he made me aware of two papers that were written quite some time ago on the subject of alternative splicing. They specifically address the question of how many different proteins can be made from a single gene. The first one was written by Douglas L. Black and published in the journal Neuron. It reviewed some recent results on alternative splicing in a specific chicken gene.
The gene, which is called cSlo, is involved in hearing. The cochlea (the organ that does the actual hearing) in the chicken’s inner ear is lined with hair cells, each of which responds most strongly to specific frequency of sound. One end of the cochlea contains hair cells that respond best to low-frequency sounds, and the other end holds the hair cells that respond best to high-frequency sounds. In between, the hair cells vary smoothly so as to respond best to the sound frequencies in between. Part of why each hair cell has its own “best frequency” to detect is a result of how electrically-charged particles are transported into and out of the cell. The proteins produced by the cSlo gene aid in that process.
Well, recent studies show that the cSlo gene uses alternative splicing to create 576 different proteins, each with a slightly different electrical response. As a result, when researchers studied the hair cells on one end of the cochlea, they found one of the proteins. When they looked at the hair cells on the other end, they found another protein. In between, they found 574 other proteins, each of which was ideal for producing a strong response to a specific sound frequency!1
In other words, the exons and introns of the cSlo gene have been set up so that each hair cell can use alternative splicing to produce a protein that is perfect for the frequencies to which it is supposed to respond most strongly! The design is so detailed that each hair cell can choose from among 576 different proteins in order to get a good response. All of this is the result of a single gene!
If you think that’s impressive, hold on to your hat! A different paper reports on a study involving a gene in the fruit fly Drosophila melanogaster. This gene, called Dscam, produces a protein that resides on the surface of certain cells in the fly’s nervous system. The authors show that the introns and exons in the gene allow for the production of 38,016 different proteins! Does the gene actually produce that many proteins? The authors can’t confirm this, but they conclude that each cell produces 14-50 different mRNAs from that gene, allowing it to have 14-50 different proteins on its surface. Because there are so many possible variants, and because each cell uses several of them, there are trillions upon trillions of different possible combinations. As a result, the authors conclude:2
…the Dscam repertoire of each cell is different from those of its neighbors, providing a potential mechanism for generating unique cell identity in the nervous system and elsewhere.
In other words, it is very possible that this single gene allows for every cell to be unique from the hundreds of millions of other cells of the exact same type!
As the biology professor with whom I was meeting told me about these two papers, I considered how their results represent yet another example of the exquisite design that you see in nature. DNA is already amazing, but to think that a single gene can produce 38,000+ different proteins is truly awe-inspiring. However, the biology professor pointed out something that I hadn’t even thought about: the implications that these results have on the process of comparing the genomes of two different species.
For example, I have written several times about the difficulty involved in comparing the human genome to the chimp genome. Depending on the study, geneticists have found that the chimp genome is 72-95% similar to the human genome (see here and here, for more information). I have also pointed out that since we only know the human genome to an accuracy of 6% and the chimpanzee genome to an accuracy of 9.5%, it is not clear how anyone can really say how similar the two genomes are. However, given what we now know about alternative splicing, it’s not clear that it matters how similar the two genomes are to one another.
Let’s just say for the sake of argument that the human genome is 95% similar to the chimpanzee genome. I seriously doubt it is, but I think that’s the highest level of similarity possible, given what we know about the two genomes right now. If many genes can produce hundreds or thousands of different proteins, then in the end, even 95% similar genomes can produce radically different sets of proteins, which would result in radically different species. Thus, given the amazing design that we see in DNA, it’s not clear that comparing genomes is the best way to judge the similarities of two different organisms.
If nothing else, these two papers show me that we have still not learned the depths of the design that exists in DNA. The more we learn about God’s creation, the more I stand in awe of what He has done!
REFERENCES
1. Douglas L. Black, “Splicing in the Inner Ear: a Familiar Tune, but What Are the Instruments?,” Neuron 20:165–168, 1998.
Return to Text
2. Guilherme Neves, Jacob Zucker, Mark Daly, and Andrew Chess, “Stochastic yet biased expression of multiple Dscam splice variants by individual cells,” Nature Genetics 36(3):240-246, 2004.
Return to Text

Does alternative splicing tend to produce functionally similar proteins, or could the same gene fragment be used for wildly different purposes that happen to need the same protein stretch?
In the first case it would be perfectly conceivable that an organism could evolve genes that use alternative splicing (since a mutation that is helpful to one is likely to be similarly helpful to other proteins doing a similar job). The more distant the task, the less likely it seems that the same mutation would be helpful.
By way of analogy, if a dictionary were compressed in the same way, one could usefully mutate the root “color” to “colour” and correct the same mistake (:P) in colouring, colourful, uncoloured, and so on.
However if the dictionary compressed at the level of “or” and that were mutated to “our”, your lourdy alteration would also ruin such wourds as “boure” and “courn”, and all four the sake of courrecting “colour”. Clearly that alteration wouldn’t survive the ferocity of grammarian selection.
This is still a relatively new area in genetics, so the short answer is that we don’t really know. However, several surveys of alternative splicing, such as this one, have shown that both effects occur. Sometimes, the different proteins made by alternative splicing are very similar. However, they are sometimes radically different so that the alternatively-spliced proteins have a completely different function from the protein produced by the non-alternatively-spliced gene. The former is more common than the latter.
Here is the problem I see with your dictionary analogy. You are talking about insertion mutations and how they affect the entire word. That’s not what alternative splicing is all about. In alternative splicing, the insertion mutation must split one useful section from another, and then there has to be a mechanism by which those useful sections can be put together in different ways. So the proper analogy, as far as I see it, would be mutations on a word like “transaction.” Insertion mutations that lead to “traXXnsXXaXXctXXion” would render the word meaningless. However, if at the same time the dictionary formed an automatic “cut-and-paste” program that could chop out the X’s and splice the segments together, then we could get the words “transaction,” “tract”, “transact,” and “action.”
I didn’t mean my analogy to explain alternative splicing, but the problems that it might pose to evolution.
i.e. If using your notation the dictionary contained unXXcolorXXingXXfulXXed for producing all sorts of variations on the word color (which are functionally all very similar) then the beneficial mutation of adding a U (unXXcolourXXingXXfulXXed) would be be beneficial to all the words involved. If that were a genetic mutation it would be selected for.
However if a single exon were used in all the very different cases where “or” appears (bXXcXXcolXXorXXeXXn) the mutation required to fix the spelling of color would break words like bore and corn. If that were a genetic mutation it would be selected against.
I understood your point, Josiah. I just didn’t think the analogy applied to alternative splicing. However, you now have an excellent analogy.
And by extension make it very difficult to evolve an improvement that would otherwise seem like it’s just a single mutation away from completion, because that mutation would be more harmful than the benefit provided.
That’s incredibly amazing! I have a quick newbie question.
RNA is formed during transcription correct? In which case there must be individual nucleotides floating around the nucleus that are used to eventually form the RNA strand during transcription. Why aren’t the introns that are cut off re-used later for more transcriptions? Only one nucleotide differs from DNA and RNA, so if they are individually cut off, can’t they be reused later?
I’m assuming that I’m missing something basic here, but I am just wondering!
Thanks for your comment, Inazuma. In answer to your question, remember that the introns are cut out of the mRNA, not the DNA. So an mRNA copy of the ENTIRE gene is made, and at that point, it is called “pre-mRNA.” Then, the introns are cutout of the pre-mRNA, the exons that are going to be used are spliced together, and the result is the final mRNA that leaves the nucleus. The introns that were cut out, and any exons that weren’t use in that particular splicing, are, indeed, recycled and used in further transcriptions.