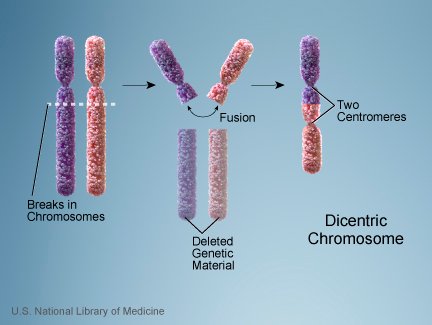
People have 23 pairs of chromosomes, for a total of 46 chromosomes. Most apes have 24 pairs of chromosomes, for a total of 48 chromosomes. One very popular piece of genetic evidence for the idea that humans and apes have a common ancestor is that human chromosome 2 looks like two chimpanzee chromosomes that have been stitched together. As the evolutionary story goes, the common ancestor between apes and humans had 24 pairs of chromosomes, and it initially passed them to those animals that began evolving into apes and humans. The apes kept that number of chromosomes, but after the human lineage split off from the chimpanzee lineage, something happened to fuse two of the chromosomes, leading to only 23 pairs of chromosomes in humans. As Dr. Francis Collins puts it:1
The fusion that occurred as we evolved from the apes has left its DNA imprint here. It is very difficult to understand this observation without postulating a common ancestor.
This idea has been around for a long time, but I never put much stock in it. Why? Because even if human chromosome 2 is the result of two independent chromosomes being fused together (an example of which is shown in the illustration above), I don’t see why this can only be understood in the context of evolution. After all, we know that chromosome fusion events happen in human beings today.2 Thus, if human chromosome 2 really is the result of a fusion of two chromosomes, it could have happened early in the history of human beings. It need not have happened to some hypothetical evolutionary ancestor. Any event that restricted the human population to those who arose from the people who originally experienced the chromosomal fusion would then fix that chromosome in the population. A worldwide Flood in which a single family was saved would be one example of such an event.
Regardless of whether or not human chromosome 2 is evidence of common ancestry, it’s getting hard to understand how it could even be the result of two chromosomes fusing.
Dr. Jeffrey P. Tomkins, a former director of the Clemson University Genomics Institute, recently published a paper in which he analyzed the genetic content of the site at which this fusion was supposed to have taken place. He found something really interesting, but to understand why it is interesting, you need to understand something about the structure of genes. In the DNA of eukaryotes (all organisms whose cells have a nucleus), genes are composed of two regions: exons and introns. When the information in a gene is copied so the cell can use it to make a protein, both the introns and the exons are copied. Before the copy is used to make the protein, however, the introns are removed, as shown in this figure:
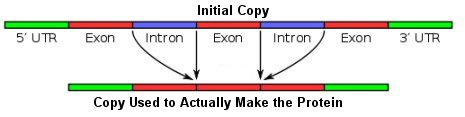
In other words, the exons are the parts of the gene that contain the information the cell needs to make a specific protein. The introns are “spacers” that separate these chunks of information. Why is a gene constructed this way? Because each exon is a module of information, and the modules can be put together in many different ways. As a result, many different proteins can be made from a single gene. This process is called alternative splicing, and it vastly increases the amount of information that DNA can store. Indeed, there is a specific human gene that can be used to make 576 different proteins thanks to the way its introns and exons are arranged.3 In the fruit fly, there is a specific gene that can be used to make 38,016 different proteins!4 This is just one of the many features of DNA that shows it is the result of incredible design.
So here’s where Dr. Tomkins’s research gets interesting: He finds that the place on chromosome 2 where the two chromosomes are supposed to have fused is right in the intron of a gene!5 The gene is charmingly named DDX11L2, and it is known to be used in several different cells, including those performing tasks related to the nervous system, muscle system, immune system, and reproductive system.
It’s really hard to understand how two chromosomes could fuse at the intron of a functional gene, but that’s not the end of the story. The fusion site also contains an important sequence of DNA called a transcription factor binding site. This is a sequence to which a molecule can attach so as to regulate how often the gene is used. So not only is the fusion site right in the middle of a functional gene, it is actually found in a region that helps to regulate how that gene is expressed. That makes it even more difficult to understand how a fusion event could have taken place there.
Is this conclusive evidence against the idea that our second chromosome is the result of two independent chromosomes being fused together? Not really. After all, the spot that Dr. Tomkins studied does bear a remarkable resemblance to the kinds of sequences found when two chromosomes fuse together. Thus, if it is not the result of chromosome fusion, we need to understand why that region of the chromosome has a sequence characteristic of such things. Also, it is well known that the human chromosome 2 has two centromeres, and typical chromosomes have only one. As the illustration at the top of this article shows, a fusion event would explain why this chromosome has two centromeres. At the same time, however, there is a known genetic process in which a chromosome can form a second centromere.6 It is certainly not a common event, but it does happen.
Dr. Tomkins’s research has definitely made it harder to believe that human chromosome 2 is the result of a fusion event, but there still needs to be a lot more research done on the issue. Nevertheless, even if it turns out that geneticists can explain how a chromosome fusion event happened in the middle of a functional gene at one of its transcription binding sites, it still offers no evidence for common ancestry. It only shows that at some point in the past, two chromosomes fused. There is no reason to think it was more likely to have happened in some hypothetical evolutionary ancestor than it was to have happened as a part of human history.
REFERENCES
1. Francis Collins, The Language of God: A Scientist Presents Evidence for Belief, Free Press 2006, p. 138
Return to Text
2. Iain D O’Neill, “Homozygosity for constitutional chromosomal rearrangements: a systematic review with reference to origin, ascertainment and phenotype,” Journal of Human Genetics 55:559-564, 2010
Return to Text
3. Douglas L. Black, “Splicing in the Inner Ear: a Familiar Tune, but What Are the Instruments?,” Neuron 20:165–168, 1998.
Return to Text
4. Guilherme Neves, Jacob Zucker, Mark Daly, and Andrew Chess, “Stochastic yet biased expression of multiple Dscam splice variants by individual cells,” Nature Genetics 36(3):240-246, 2004.
Return to Text
5. Jeffrey P. Tomkins, “Alleged Human Chromosome 2 ‘Fusion Site’ Encodes an Active DNA Binding Domain Inside a Complex and Highly Expressed Gene—Negating Fusion,” Answers Research Journal 6:367-375, 2013 (available online)
Return to Text
6. Warburton PE, “Chromosomal dynamics of human neocentromere formation,” Chromosome Research 12(6):617-26, 2004
Return to Text

If the chromosome 2 fusion event never happened then humans would have 48 chromosomes. Wouldn’t that put us on par with the apes genetically? Are there other animals that have the same chromosome count but look vastly different or are they the same? Is it possible that the fusion is what makes us human instead of apes?
Chromosome count has nothing to do with genetic similarity, Luis. For example, African wild dogs, domestic chickens, and doves all have 78 chromosomes. No one suggests that African wild dogs, domestic chickens, and doves are genetically on par with one another. The American beaver, European polecat, hyena, mouse, mango, and ferret all have 40 chromosomes. Once again, no one is suggesting that they are genetically on par with one another. We are incredibly different from apes on a genetic level, regardless of the closeness of our chromosome count.
I am blown away by the complexity of our bodies and that of all living things. Just to think that scientist can determine these kinds of details is really amazing. Unfortunately it is not commonly taught or described. I think if most science books spent more time on real science instead of focusing many chapters to theories, ages/time periods, etc. more students would be excited by science. I appreciate your books and blog.
Thanks, Lindy. I am glad my books and blog are valuable to you!
Something that I have always found interesting is that chromosome count does not seem to correlate with complexity.
Since chromosomes store genes and those genes store information. One would think that the more chromosomes that an organism has would lead to it being more complex. It is true that more chromosomes means more information storage but it does not mean more complexity.
For instance humans have 46 chromosomes and the Adders-Tongue fern has 1260 chromosomes which means that the fern potentially has enough information to describe us 27 times. Yet I would consider humans far more complex.
Perhaps the Intron Exon relationship in the fern’s chromosomes yields a much lower combination of proteins then our own. Or maybe the size of the fern’s chromosomes are much smaller than human chromosomes leading to an overall smaller number of genes.
Either way it is still all very interesting.
It is very fascinating, Aaron. It is certainly true that chromosome count has nothing to do with an organism’s complexity, but you do have to be careful when talking about plants. For reasons we don’t understand, plants go through whole-genome duplication events rather easily. As a result some plants have an absurd number of chromosomes, most of which are either deactivated or strongly down-regulated.
So are you saying that plants duplicate their entire genome and therefore have redundant genes? And since they have these duplicates that make the same proteins they compensate by producing inhibiters that allow the genes to be expressed at the same ratio as before the duplication?
I would not be surprised if this is the case since the entire transcription factor process seems to be highly regulated by feedback loops.
The real question seems to be why the duplication happens in the first place. Is it yet another form of data redundancy to protects against mutation?
I am not very knowledgeable on this issue, Aaron. Here is a good article that discusses the advantages and disadvantages related to it.
“As a result some plants have an absurd number of chromosomes, most of which are either deactivated or strongly down-regulated.”
Do you mean that they are full of junk DNA?
Luis, downregulated DNA is not junk DNA. Junk DNA is DNA that has degraded to the point that it can’t have any function. Since we know the vast majority of an organism’s genome is functional, multiple copies of that DNA are also functional. It might not be used, but it is functional. Saying unused DNA is junk DNA is like saying that because I don’t use something, it is junk. So if a person has an Aston Martin in mint condition and chooses not to drive it, is it a junk car? Of course not.
I love how one answer can lead to hundreds of new questions. As my father says “The more you learn the less you know”.
I am fine with that because I believe that we are destined to spend the rest of eternity trying to figure out what God did in the first six days.
I wholeheartedly agree, Aaron!
…Reading through the comments on these recent posts….Love this blog/ministry!
I have an OT question that I was looking to post on a more related topic, like your review of Gerald Schroeder’s book, but comments were closed there. Does General Relativity explain distant starlight? And if so, how? Schroder gets around this by “frame of reference” so it’s both at the same time. But from the human perspective (location and velocity in the Universe) starlight is light years off, correct? Or does it relate to when we were created, relative to the rest of the physical creation in Genesis. I halfway understand all of this, so I might not have phrased correctly, but do get the general ideas.
So, just wondering if you had an opinion or could direct me to a link, Dr Wile? Can’t say I would understand, but certainly would like to try. Thank you so much!
That’s a great question, Elizabeth. WordPress (the software that runs this blog) turns comments off after a while, so I am happy to answer your question here. As you suggested, General Relativity does answer the distant starlight question. Since larger gravitational fields slow down the passage of time, in order to determine how long it took light to travel from a distant part of the universe to here, you need some model of how gravitational fields vary throughout the universe. Those who believe in the big bang make the absurd assumption that on a large enough scale, the universe is homogeneous. Of course, that assumption goes against nearly every observation that has ever been made. However, if you make that absurd assumption, then you can treat time as passing at the same rate pretty much everywhere.
In his book, Starlight and Time, Dr. Russel Humphreys makes a much more reasonable assumption. He assumes that the universe expands as a sphere, which is utterly different from how the big bang views the expansion of the universe. Using Dr. Humphreys’s more reasonable assumption, General Relativity tells us that time passes more quickly the closer you are to the center, and it passes more slowly the closer you are to the edge. Thus, if the Milky Way (the galaxy in which we reside) is close to the center of the universe, thousands of years will have passed on earth, while billions of year will have passed near the edges of the universe. As a result, light would have no trouble reaching us, even from the edge of the universe.
Here is a basic discussion of Humphreys’s view. Here is a variant of that view, which I think probably has more merit.
That’s so cool! Will look into that book/explanation…makes so much sense from the science. Thank you for your answer, Dr. Wile. Merry Christmas!
Your on my RSS feed now. Thanks for writing.
My background is in church history and education, so this is pretty much beyond me, but I appreciate your attempt to explain it in terms I can understand.
Knowing that your philosophical worldview will not affect the conclusion of your work (as is the case with evolutionary thought) is comforting and motivates me to keep reading.
Thanks again.
Jonathan, I wouldn’t say that my philosophical worldview doesn’t affect the conclusions of my work. All scientists are biased, including me. I just try to be as open as possible to the data and let them guide me in making my conclusions. That’s what led me to Christianity, so I have no reason to stop doing that now!
Hi Dr. Wile,
I was a bit disappointed that Dr. Tomkins made the leap that chromosome fusion has been thoroughly refuted, which as you indicated is not necessarily the case. It appears that the DDX11L family of genes are typically only found in subtelomeric sequences on the ends of chromosomes, and its presence at the fusion site would seem to support the idea that an end-to-end telomere fusion occurred. See Costa et al. (2009), http://link.springer.com/article/10.1186%2F1471-2164-10-250/fulltext.html. According to that article, which is on the subtelomeric regions of humans and does not specifically talk about DDX11L2, “…the human DDX11L gene family members showed a significant sequence similarity with the DDX11 homologous gene of chimpanzee and rhesus on chromosomes IIp and 11, respectively.” I don’t know where the DDX11 on chimpanzee chromosome 2p is found exactly, but I presume it is in the subtelomeric region. I found this interesting, so looking back through Dr. Tomkins’ paper I was surprised to see that he did actually cite this work by Costa and company multiple times. In one instance, he summarized Costa as follows:
“Costa et al. (2009), reported that at least 18 different DDX11L-like genes exist in the human genome. They also reported that very little synteny existed for these genes in apes. Using fluorescent in situ hybridization in chimpanzees and gorillas, only two locations of similarity for DDX11L-like genes were found in chimpanzee and four in gorilla—none of which corresponded to locations in human, or each other in apes. Of key importance to the topic of this paper was the fact that none of the regions of DDX11L hybridization in the chimp or gorilla genomes occurred on chromosomes 2A or 2B.”
This characterization of the paper seems to contradict the quote from it that I gave above, where a very similar sequence exists on 2p, one of the pair that is claimed to have fused in humans. I’ll keep reading and respond here if I find anything wrong with what I’ve said.
Costa et al.:
“The DDX11L gene family members derive from a rearranged portion of the primate DDX11 gene (alias CHLR1, homologous to CHL helicase of Saccaromyces Cerevisiae), propagated among many subtelomeric locations (see table 1) as part of a segmental duplication [18]. The human subtelomeric DDX11L genes show up to 98.5% sequence identity with the exons 18 and 22–25, and the 3′ UTR of DDX11 gene. The finding that DDX11L is a novel multicopy gene family present in different human and primate subtelomeres, and that all the identified gene copies undergo canonical splicing mechanism – and are also transcribed (at least in humans) – suggests this gene is emerging from an inactive pseudogene, and is probably undergoing a neo-functionalization process [18].”
That is a mouthful 🙂 But I thought it summarized the situation well.
Thanks for that helpful comment, Jon. I have not read the Costa paper, so I can’t comment on why Dr. Tomkins seems to be saying something different from what the paper says. However, I do agree that the situation is far from settled, and Dr. Tomkins is not being as careful as he should be in saying otherwise.
This is why I don’t trust creationist science. This is proof that I was right about creationists twisting the science and the quotes to match their beliefs. Dr. Wile, you gave me a link to Tomkins work concerning the 70% human-chimp similarity. Are you going to tell me that you trust his results after what you just claimed about him not being careful? How many of the creationist links that supposedly show the reliability of Genesis you gave me should I trust?
Luis, once again you seem to confuse the scientist with the science. I can give you example after example of evolutionists twisting the science to match their beliefs (see here, here, here, here, and here for just a few examples). Does that make you distrust evolutionary science? You should not trust any scientist. You should trust only the evidence. Besides, Dr. Tomkins isn’t the only one who has demonstrated that the human and chimp genomes are only about 70% similar. As the article I gave you discusses, a completely different scientist did a completely different kind of analysis and came up with roughly the same number.
Also, how do you know that Tomkins twisted anything? I have not read the article Jon quotes from, but as he indicates, the statements are incredibly technical. It is very possible that Tomkins is not twisting anything but instead is simply using a different interpretation. Can you tell me exactly how Tomkins twisted the science? If not, you shouldn’t accuse him of doing so! Are you saying we shouldn’t trust any work from Tomkins because he wasn’t very careful in one of his papers? If so, then you’ll have to distrust the work of most scientists, because most scientists show a lack of care in their analyses from time to time, especially when the data seem to support one of their pet ideas (see here, here, here, and here, for example)! That’s part of being human.
Hello Dr. Wile!
I found a link going to a response to this article on some Biology Answers sort of page. What do you think?
http://biology.stackexchange.com/questions/5558/chromosome-2-fusion
Thanks for the link, Alex, but the discussion is not about this study. It is about two earlier papers by Bergman and Tomkins regarding the nature of the human chromosome and how it argues against chromosome fusion. I tend to agree with the responder, that evolutionists have reasonable explanations for why aspects of the structure of human chromosome 2 don’t look like what one would expect from a fusion event. For example, as the responder says, you would expect repetitive DNA around a now-defunct centromere to be excised quickly, so it’s not surprising the supposedly-defunct centromere doesn’t look much like one and contains few telomere motifs.
I think the study covered in this post adds a very significant point to which I can’t imagine a good answer: how is it that fusion occurred at an intron of a functional gene, especially an intron that contains a transcription factor binding site? Now of course, just because I can’t imagine a good explanation doesn’t mean there isn’t one. That’s why I agree with Jon that this is not a slam-dunk case like Tomkins thinks.
Hey Luis,
Every explanation for facts is a different “twist”, so I don’t think Dr. Tomkins is being dishonest. My issue was that he is not being as conservative as he should be for a scientific paper, but these types of less-rigorous claims are frequent in common speech. I see this in papers in all fields, unfortunately.
I was not addressing the central argument of his paper in my first post, but I will look at it here. And I did miss something regarding the apparent contradiction between what he said and what the Costa paper presented, which I will explain here as well.
Disclaimer: Unlike the individuals writing these two papers I am not a geneticist, and I have not been trained in reading this type of paper nor the techniques involved. The Costa paper especially is very jargon-heavy, and after several reads I am just beginning to understand it.
There is a gene family called DDX11L, and around 18 instances of it are found in the human genome. All of the known instances are found at the ENDS of chromosomes, in the “subtelomere” regions, except for a single gene in this family: DDX11L2. This one is right in the middle of chromosome 2, at the site of the potential fusion. As Costa et al. reads, “we defined a novel gene family – the most telomeric ever identified – named DDX11L.” That is why I consider its placement as a piece of evidence for the fusion theory. BUT, it also raises an apparent problem for the fusion as well, which I will get to.
It was known since 2002 that the fusion site was in the middle of a pseudogene. Tomkins wrote his paper in response to new information about this gene, which is now called DDX11L2. He says: “The location of the putative fusion sequence inside a functional and highly expressed gene associated with a wide variety of cellular processes strongly negates the idea that it is the by-product of a hypothetical head-to-head telomeric fusion.”
Costa says, “Although we have not yet defined a biological function for the newly identified transcript family, we found that these genomic regions are actively transcribed (in many human tissues) and also undergo canonical splicing. To date, we cannot define these transcriptionally active regions as functional genes, but we cannot definitely exclude it.” The Tomkins paper of course asserts the gene is functional. Tomkins here again may be jumping the gun in saying that this gene has a useful purpose, but he points out that it is expressed in the same tissues as DDX11 is. I don’t know if its utility is really significant to the issue, however. There is still the question of how the gene ended up straddling the fusion site in the first place! Because it is a telometric sequence, it seems unlikely that it moved there AFTER the fusion. If two chromosomes fused, it must have been carried there on the ends of one of the original chromosomes, but I don’t know how it could then be overlapping the fused region across BOTH original chromosomes.
The subtelomere regions are highly variable compared to the interior sequences, which is the basic subject of the Costa paper. Interestingly, in the three humans that were tested, at least one person had the DDX11L gene at the END of chromosome 2 instead of at the fusion site. “The human DDX11L genes are at high risk for genomic rearrangements, such as deletions or translocations. Thus, since we have detected polymorphic rearrangements by the analysis of only three unrelated human individuals, on three different chromosomes, we cannot exclude that other intact or partial copies of DDX11L genes might be identified within telomeres of other individuals.” I wonder if this has something to say about whether the gene is functional or not? I don’t know if a functional gene can be moved around like that, without harming the person. If not, the fact that it relocated would be evidence that it is a nonfunctional pseudogene at the fusion site. (It looks like it is possible through a multi-step process for a functional gene to relocate–but I could be wrong on this.) Regardless it seems most likely that it started out in the midst of the fusion site rather than starting out at the end and coming inward, so a relocation probably can’t explain the current placement.
The researchers used a technique called “FISH analysis” (fluorescent in situ hybridization), where fluorescent probes tailored to match a certain gene sequence go out and bind to those chromosomal sequences they match up with, taking advantage of the DNA strand binding (“hybridization”) that occurs during sexual reproduction. You can see in the Figure 3 image where the matching sequences were found. I recommend you check out Figure 3, the picture really is worth a thousand words (again the paper address is http://link.springer.com/article/10.1186%2F1471-2164-10-250/fulltext.html). Compare the two images of chromosome 2 on the leftmost side.
I pointed out an apparent contradiction before between the two papers, where Costa claimed that the DDX11L family has a significant similarity to the DDX11 gene on chimpanzee chromosome 2q, and then Tomkins cites Costa to claim that no DDX11L-like gene was found in apes that corresponded to the one on chromosome 2. It turns out that Tomkins was referring to the “FISH analysis” results, which did not, in fact, detect a similar gene on chimpanzee chromosome 2q. And of course, DDX11 is a different type of (similar) gene. I think Tomkins was perhaps justified, then, in his comments on this particular point.
I think the presence of a subtelomere gene at the potential fusion site of chromosome 2 is very interesting. But, that being said, I’m not sure what it implies that this gene crosses the fusion site, and that it is found at the end of chromosome 2 in some humans.
This post is a long-winded way of saying “I don’t have the expertise to know what to make of these facts.” 🙂
Thanks for your take on the study, Jon. I agree that it is very unlikely for the gene to have moved, so you have hit the nail on the head. If this is the result of fusion, the gene had to have been carried at the ends of both chromosomes, and somehow, the fusion had to happen just right so as to preserve the gene. I agree that the amount of functionality is not all that important. If the gene is expressed at all, it seems to me that it is hard to explain how the fusion event could have happened so as to preserve the cell’s ability to transcribe the gene in a regulated fashion.
You have also hit the nail on the head with regards to Tomkins’s conclusions. The facts of the study seem completely reliable. The problem is simply that he is not cautious enough in his conclusions. As you say, however, this happens throughout the scientific literature, so while it is regrettable, it is not unusual.