(Copyright Answers in Genesis, published at http://www.answersingenesis.org/articles/arj/v6/n1/human-chimp-chromosome in a study by Jeffrey P. Tomkins)
PLEASE NOTE: The results of this study are known to be wrong due to a bug in the computer program used. A new study that uses several different computer programs shows an 88% overall similarity.
I have written about the similarity between human and chimpanzee DNA three times before (here, here, and here). It’s an important question for creationists, intelligent design advocates, and evolutionists alike, since the chimpanzee is supposed to be the closest living relative to human beings. As a result, a comparison of chimp DNA to human DNA gives us some idea of what the process of evolution would have to accomplish to turn a single apelike ancestor into two remarkably different species like chimpanzees and people.
Early on, it was widely thought that human DNA and chimp DNA were 99% similar. As I discussed in my first post on this subject, that was based on a very limited analysis of only a minute fraction of human and chimp DNA. Now that the entire set of nuclear DNA (collectively called the “genome”) of both humans and chimpanzees have been sequenced, we now know that the 99% number is just plain wrong. Interestingly enough, however, even though both genomes have been fully sequenced with a reasonable amount of accuracy, no one can agree on exactly how similar the two genomes are.
Why is that? Because comparing genomes is a lot harder than you might think. While we know the sequence of the chimp and human genomes really well, we don’t understand the DNA itself. Indeed, there are large sections of DNA that seem to be functional, but we simply have no idea what they do. As a result, comparing the genomes of two different species can be very, very tricky.
Probably one of the best explanations of just how tricky DNA comparison is comes from Dr. Richard Buggs, a geneticist at Queen Mary, University of London. Back in 2008, he wrote about the steps he would take to compare the human and chimp genomes, and if you read his explanation, you will get an idea of how difficult such a comparison is. His conclusion was:
Therefore the total similarity of the genomes could be below 70%.
Since that time, the chimpanzee genome has been sequenced to an even better degree, and other methods have been used in an attempt to determine the similarity between the chimpanzee and human genomes. One of the more popular methods is based on an algorithm called BLAST, which chops up DNA (or proteins) into small segments and then tries to compare them to the segments on a different set of DNA (or proteins). This seems like the most “generous” way to compare two genomes, because it doesn’t require one genome to be structured similarly to the other. The only thing that matters is whether a bit of information in one genome can be found anywhere in the other genome.
Using this method to determine the similarity between the human and chimp genome, researchers have come up with different answers. Dr. Todd Wood, an expert in genome comparison and former Director of Bioinformatics at the Clemson University Genomics Institute, did a BLAST analysis that indicated human and chimp DNA are roughly 95% similar. However, Dr. Jeffrey P. Tomkins, former director of the Clemson University Genomics Institute, did a different BLAST analysis and concluded that the similarity was 86-89%.
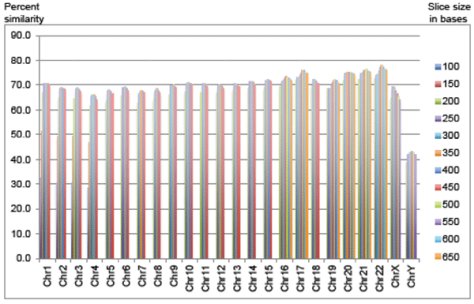
Well, Dr. Tomkins just published a new study, and as far as I can tell, it makes the most sense of any BLAST analysis done so far. In this study, he chopped up the chimpanzee genome into “slices” that were as small as 100 base pairs long or as large as 650 base pairs long. The chimpanzee genome is 2.9-3.3 billion base pairs long, so obviously these slices are incredibly small compared to the entire genome. He then looked for each “slice” on the human chromosome that is supposed to correspond to the chimp chromosome where the slice was found. The two slices didn’t have to match exactly; they just had to be similar enough to think that they could be related to each other.
The graph at the top of this post shows his results. Notice that the similarity hovers around 70% for all chromosomes except the Y chromosome. The size of the “slice” affects the result a bit, but really not much. In the end, this leads Dr. Tomkins to conclude:
Genome-wide, only 70% of the chimpanzee DNA was similar to human under the most optimal sequence-slice conditions. While chimpanzees and humans share many localized protein-coding regions of high similarity, the overall extreme discontinuity between the two genomes defies evolutionary timescales and dogmatic presuppositions about a common ancestor.
Is this the last word on the subject? Most certainly not. I think it is probably the best comparison attempt made so far. Also, the fact that the Y chromosome has a remarkably low level of similarity compared to the other chromosomes is consistent with another study. In addition, the results essentially agree with Dr. Buggs’s analysis, which was based on a completely different strategy. At the same time, however, there is a huge discrepancy between this analysis and Dr. Wood’s analysis. In addition, as we learn more about genomes and how they work, we will probably find better ways to compare the genomes of different organisms.
For right now, however, it seems clear that humans and chimpanzees are not nearly as genetically similar as most evolutionists would have us believe.

I remember an article by Answers in Genesis that pointed out that even in the most optimistic of conditions in the similarity of human and chimp DNA (99%), the 1% difference is so drastic on a molecular scale that the odds are sharply against chimp-to-human evolution. I can’t recall what specifically made the 1% difference so great, but it goes to show that evolution doesn’t have a chance.
For me, I think you can just point out the flaws of abiogenesis and the debate is over, but I like to see the evidence continually breaking down the macroevolutionary hypothesis.
Thanks for the comment, Jacob. The article might have been about Haldane’s Dilemma, which indicates that even using wildly unrealistic assumptions that would make evolution as fast as possible, in 10 million years, only about 0.02% of a genetic change could occur from the supposed common ancestor of humans and apes. Thus, even a 1% change would be considered far too much for evolution to accomplish. Of course, evolutionists have ways of explaining around the dilemma, but they don’t seem to be consistent with what we know about genetics.
I agree that the origin of life is the biggest hurdle to any form of materialism. That is why many evolutionists try to strike a very big contrast between evolution and abiogenesis.
Here is what would make this more interesting — do the same comparisons on a number of creatures. If the difference between a mouse and a human using this technique is also around 70% then this is very interesting. If it is, say, 30% or something, then it would actually point towards a macro evolutionary picture.
But beyond that, how you compare things is pretty tricky. For example, how would you compare the KJV and the NIV bibles in a way that confirms they come from the same source? Might result in a 5% or 99% measurement depending on your technique.
So in the end I think this is a totally non-interesting result. Except to point out that whenever you hear a really stereotypical number like “99%” you should probably ask questions.
I am probably going to have to disagree with you, Skip. Even as a creationist, I would expect human and mouse DNA to be more different than human and chimp DNA. I would expect human and fish DNA to be even more different. That’s because the biochemical needs of humans are closest to the biochemical needs of a chimp, more different from the biochemical needs of a mouse, and even more different from the biochemical needs of a fish.
Also, I think this is a very interesting number. If human and chimp DNA really are only 70% similar, that gives us an idea of what enormous work evolution had to accomplish. It had to take some hypothetical common ancestor’s genome and rework that genome significantly to come up with both chimps and people. This puts a severe constraint on the mechanism by which evolution could have accomplished such a heroic feat.
Now where I will agree with you is on your analogy to the Bible. If we compared the KJV and NIV on a word-for-word basis, they would look incredibly different. However, if we converted each word to a definition and then compared them on a “word definition” by “word definition” basis, they would come out a lot more similar. If we then converted each sentence to its meaning and then compared them on a “sentence meaning” by “sentence meaning” basis, they would look even more similar. Hopefully, as we learn more about how DNA actually works, we might be able to use similar tactics to compare genomes. I suspect that would be a lot more illuminating than just comparing nucleotide base sequences.
An interesting article Dr Wile, thank you.
Please allow me to share an article I came across while on a Darwin vs Design course.
The Chimp-Human 1% Difference: A Useful Lie 06/29/2007 (Report by David Coppedge)
https://docs.google.com/document/d/17PTD6sdmtYw3nhszb16dE6-W23UhkBiY1mS4sB4PReo/edit?usp=sharing
I think Skip has a point but looking at human and mouse wouldnt’ really be informative because of homoplastic variation. However, as I have been pointed out on many occasions elsewhere, what would be a better comparison would be looking at the differences between genomes of species that creationists believe share a common ancestor by virtue of their being the same “kind”. If one applies the same BLAST methods Tompkins did for Chimp/Human to say sheep and goats or cheetahs and leopards or foxes and wolves what would one find? If these are 70% different what does that tell us about these species and does the 70% similarity between humans take on a new meaning? I think that creationists need to be careful about saying that they are sure that genetic similarity can be used as a proxy for the delineation of kinds. I would predict that foxes and dogs are at least 30% different from one another using the same methods of estimating differences and yet AIG and others are fairly quick to claim that foxes and wolves arose from a single pair of ancestors.
That’s an excellent idea, Joel. As I understand it, both the cow genome and the yak genome have been sequenced. Creationists think they are part of the same monobaramin, so it would be very interesting to see how a BLAST comparison of those two genomes works out. I will suggest this to Dr. Tomkins. I would predict that the genomes would turn out to be very similar, but I do agree with you that creationists (and everyone else) need to be careful in connecting genetic similarity (especially when it comes to just the sequence of nucleotide bases) to common ancestry. We just don’t know enough about DNA yet.
unfortunately where this was published will for the time being make it easy for people to disregard.
You are right, Antonio, and it is very unfortunate.
The whole comparison of genes to prove ansestery is flawed in the first place. just because dna is similar doesn’t nessisarily mean 2 animals are related. take the dolphin and the bat. in more than 1 aspect, their dna is remarkably similar. now, which evolved from what? the dophlin from the bat, or the bat from the dolphin?
I don’t think it’s a flawed concept, Caden. We know that evolution happens – even young-earth creationists depend on evolution to turn the animals on the ark into all the diversity of animal life we see today. Since evolution has to happen at the genetic level, by comparing genomes, you should be able to learn whether or not two animals are related by common ancestry. Now…we don’t understand DNA well enough to do that, but even our limited comparison of genomes should tell us something about which organisms are related by common ancestry and which are not.
In your dolphin/bat example, you are correct that in some parts of their DNA, they are remarkably similar. However, that’s why you have to look at the entire genome. The little brown bat’s genome has been sequenced, as has the bottlenosed dolphin’s. Unfortunately, I have not seen any comparison between the two on a genome-wide scale. I suspect that if such a comparison were made, it would show that overall, the genomes are rather different.
Now I will say this: no evolutionist would believe that the genetic similarities between a dolphin and bat are the result of having inherited those genes from a common ancestor. Thus, they have to assume that virtually identical genes (such as the ones used in echolocation) evolved independently in two radically different ecosystems. That, of course, is rather hard to believe. When I see “genetic modules” that appear in radically different creatures, that leads me to conclude the creatures were designed, because one thing we know about designed systems is that they often include subsystems that were designed for other applications.
Hi Caden,genome comparisons are indeed difficult. There are always going to be genes that are very similar even between very different organisms because of constraints on their function (and thus they must have particular amino acids in a particular order at the binding sites). The evolutionists will say these sequences are constrained through common descent the creationists through common design. There are other regions that will vary wildly sometimes even between individuals. It can be fairly easy to cherry pick sequences to make something look either similar or different. BTW, you have promoted a common misconception of evolution when you said “which evolved from what” Evolutionary theory says that neither evolved from the other but both evolved from a common ancestor that was neither a bat nor a dolphin. I was looking at a “phylogeny” of cats that AIG posted on their site that shows a “generic” cat looking thing that then evolved into lions, tigers, domestic cats. They actually have the right idea there: lions did not change into tigers or into domesticated cats but a common ancestor that was neither a lion or tiger or a house cat. Now the question is did they do that by simply resorting variation or did they evolve new genes. Genetic genome comparisons will help us sort that out. I believe the differences between a lion and cheetah or domestic cat will be much greater than that between a chimpanzee and human and so I think it is a stretch for AIG to claim that this “sorting” out of genes from a common ancestor kind can explain such incredible genetic divergence.
Hi Joel, is it actually true that they MUST have particular amino acids in a particular order at the binding sites? The variation in the way DNA letters can code for amino acids seems rather wide, and I would think, just as there are plenty of ways to write a similar computer program, there should be plenty of ways to write a sonar program. So coding options, at least to my current understanding, are quite vast. I am not a biologist, so I could be VERY WRONG on this. These two creatures, the dolphin and the bat, are radically different in most of their biological function, other than that they both use sonar, use clicks to locate prey. They are on complete opposite ends of the spectrum. My guess is that the common ancestor for a bat, should look somewhat like a bat, just like the common ancestor for a dolphin, should look somewhat like a dolphin. The common ancestor of humans and chimps would not look like a bird, nor a bat, it would look very much like a human or a chimp. With cats, of course the common ancestor is going to look somewhat like the cats, have similar functions to the cats, so I’m not sure how the argument really applies to bats and dolphins based upon the actual standards of what a common ancestor can or cannot be. The whole point of the research by AIG was to make sure that they did NOT cherry pick and provided as objective of a search as possible at this time and with our current understanding, so they should be commended for their objectivity. I am personally with Dr Wile on this subject, I believe that the sequence, if reported the same way this genome was sequenced, will give very similar results for Yak and Cow, I’d predict upwards in the 90-99 percent similarity between the Yak and Cow because they both have VERY similar biological functions, birthing processes, mental processes, lifestlye, etc. Only time and research will be able to tell though. I do agree that not enough about genomes is known to say that difference in genomes automatically means they couldn’t have an evolutionary origin through a common ancestor. It should be noted that the reason that evolutionists originally said that the genomes are 99% similar is because such changes from said hypothetical ancestor would not be quite as radical, would not take as much time to diverge from that common ancestor into the separate kinds. So the falsification of the prediction by evolutionists that the genomes between apes and humans should be nearly identical is important to the advancement of what is the truth on this matter, one which further research should help clarify.
Hi Eric, I probably wasn’t clear about the amino acid thing. I was thinking of some genes where there are important amino acids at specific points in the 3D structure of a protein. For example, in the hormone leptin (OB gene)there are are a series of positions in the protein for which all mammals have the exact same amino acids. Although there is redundancy in the genetic code for some amino acids the DNA sequence is still constrained to some very high similarity for portions of the gene. Overall similarity is hard to really relate to because even in a single gene there is going to be regions of very high similarity even among families while some areas could be highly variable between individuals. The average is just that, an average.
Regarding the YAK/cow comparison. If you mean an comparison like Tomkins did for humans/chimps 99% would be shocking. My guess would be using the same techniques probably more like 80-85% at best. But if you are talking about just gene coding sequence then definitely 99% similar (but chimps/humans are 98 or maybe 97% for coding sequence only). I just did a quick comparison for fun. I looked up the Yak leptin gene sequence and aligned it with the cow leptin sequence. They are 98% similar over the 3576 bases I aligned. There are exactly 60 individual base differences between the two and 7 insertion/deletion mutations. Now, some of these sequence is a intron (spacer between sections of coding sequence) and most of the differences are in this section. It looks like the coding sequence is 99+% the same and the intron is about 95% the same. I am sure that despite these difference that the leptin gene works the same in both animals but those 60 difference all came from somewhere if these two had a common ancestor. This gene is very “conserved” and so I would expect that other genes would show more variation than this one so probably 98-99% similar for coding sequence and maybe 94-96% similar for the rest of the sequence. But like I said before, if we include repetitive DNA, and large changes in spacer sequence sizes like Tomkins then I am sure that the similarity number will fall to the 80s at best.
Let me just jump into the conversation to say two things:
1. Tomkins says that he will look into the yak/cow comparison. However, he says that in order to do the same kind of comparison, the genomes would have to be well assembled. He says that many animal genomes are sequenced but not well assembled, so he will have to look into the status of those two genomes.
2. I would be incredibly surprised if the cow and yak genomes are only about 80% similar. Hopefully, someone will eventually be able to do that study or a similar study so we don’t have to speculate.
Thanks, I am sure that would be an interesting comparison. There are plenty of well assembled sequences that he can play with if the Yak isn’t good enough. I think buffalo has been sequenced multiple times and buffalo and yaks are pretty similar. Also, there are many dog breed sequences and even the Neanderthal sequence might be good enough now. There are also multiple rat species genomes and the mouse.
I just looked at 12 different genes in the YAK and compared them to Bos taurus(cow). They all align quite well with almost no gaps. Almost every gene has a 2% sequence divergence. These are all protein coding genes and so this represents the the portion of the genome one would expect to have the highest sequence similarity. So I would say the starting point is a minimum of 2% difference. Translated over 2 billion bases that would be 40 million difference in the DNA sequence. sounds like a lot but really not that much considering the vast majority would have no effect on phenotype.
I’m only a layman, but this bit from Dr. Thompkin’s study made me wonder:
“For the chimp autosomes, the amount of optimally aligned DNA sequence provided similarities between 66% and 76%”
If a sequence has 999 nucleotides the same but one different, is it no longer optimally aligned and the entire sequence counted by Thompkins as “unaligned”? If someone with more background in genetics could weigh in, I would really appreciate it.
Thanks for the question, Joe. That’s not what “optimally aligned” means. Remember, Dr. Tomkins took a “slice” of chimpanzee DNA that is 100-650 base pairs long and compared it to the sequences found on an entire human chromosome. Obviously, there will be more than one region of the human chromosome that is similar to that slice of chimpanzee DNA. Thus, he looks for the part of the human chromosome that is most similar to the slice of chimpanzee DNA he is considering. He then says that this slice is “optimally aligned” to that section of the human chromosome.
As an example, let’s suppose I have a slice of chimp DNA that is 400 base pairs long. When I do the analysis, I find one section of 400 base pairs on the corresponding human chromosome where 360 of the base pairs are identical but 40 are different. As I continue to search the rest of the human chromosome, however, I eventually come across a second sequence in which 375 base pairs are identical and 25 are different. I would say that the second sequence on the human chromosome optimally aligns with the slice of DNA I am considering, and at this optimal alignment, the sequences are 375/400×100 = 93.75% similar.
Thanks Dr. Wile! That makes more sense than how I was reading it.
Interesting, btw I assume that’s just a math error above as that looks like 93.75% to me:-) I’m still trying to make sense of Tomkins methods. I’ve read the materials and methods over and over but not sure what it means, but bioinformatic isnt’ exactly my strong suit. Looking at Buggs and Tompkins I can see that optimally aligned means where it best matches but then there is no criteria given for sequence similarity of the individual fragments for to know is there is a cutoff point for alignment. My overall impression is that what they are measuring is genome architecture rather than similarity of code itself. Of course similarity of architecture is a very legitimate question. Small fragments are needed to get optimal alignments because if you go to big fragments the genomes have so many gaps, insertions and rearangments that large fragments produce no good alignment. 400 bp fragments are needed to find matching (whatever matching means) sequences. In the end, I still wonder, what does 70% mean? Without a point to compare to its a bit like have one data point on a graph and wondering how to draw the line through it. Since 30+ percent of the genome are long and short interspersed repeats/copies that introduces huge amount of variation in the structure of the genome and I can’t figure out how this analysis might be effected by those elements.
As an aside, I tried doing a BLAST of several 50,000bp or larger fragments of YAKs against cows and none of them would align significant portions because it looks like there are too many insertions/deletions and inversions that get in the way. It also appears that there was just a publication that details the YAK genome compared to cows and they found numbers close to what I predicted. In the supplemental docs they actually compare YAK/Cow to Human/Chimp and the comparisons are remarkably similar in terms of total differences in protein coding genes. http://www.nature.com/ng/journal/v44/n8/extref/ng.2343-S1.pdf This is why I continue to point out that looking for total genetic distance isn’t going to define being a human. here is the Nature paper that I think is open access: http://www.nature.com/ng/journal/v44/n8/full/ng.2343.html Note that Yaks are reported to have 170 genes not found in Cows (meaning that no gene with enough similarity has been located or the location were the gene is in one genome is clearly missing in the other). Likewise cows have a similar number of genes not found in Yaks so there is some significant genetic departures between these genomes.
Yes, Joel, that was a math error. I fixed it. I agree that without other data points, a number of 70% doesn’t mean as much as it would with some more data points. However, it does mean, as I state at the end of my post, that human and chimpanzee genomes are not nearly as similar as what many evolutionists want us to believe.
The article you linked is unfortunately not very applicable to the discussion. The only comparisons I see deal with genes. While this is interesting, it has little bearing on whole-genome comparisons, since genes make up such a tiny percentage of the entire genome. However, if we do limit our discussion to genes, I don’t see how you come up with the idea that the comparisons are remarkably similar. They are quite different. Look, for example, at Supplementary Figure 10. In the case of the yak and cow (graphs A) the number of genes that are highly similar is significantly greater than the number of highly similar genes for the human and chimp (graphs B). Indeed, the number of orthologous proteins is greater across the board for the yak and cow as compared to the human and chimp. So this analysis seems to say that even when you just restrict yourself to looking at genes, the yak and cow are significantly more similar than the human and chimp.
Hey Joel, Thanks for the clarification on that. You explained it quite well. I’m not a biologists or geneticist, so it always helps to understand the specifics of these things so I can get my mind around the results and what they may indicate or not indicate at this point. That would make a lot of sense that the 3-d structure for the hormone Leptin would be identical in all mammals since all mammals need to know when to eat and when not to eat and how much to eat, or they will get either too fat or too small and perish. But I would say that it makes more sense that they would be that way if they were commonly designed. Putting my self into an evolutionists shoes, I would find it highly unlikely that the exact same or very similar sequence for a mouse, stood almost unchanged with the vast load of mutations, as well as common ancestors, that have happened over time. The thing that doesn’t make sense is that, according to evolution, the mutations in the genome are exactly what is supposed to produce the structure in the genome that codes for the hormone to begin with. Now at this point, you could say natural selection has conserved, DNA repair mechanisms have conserved, etc, but we know that before the 3-d protein coding for Leptin was created through genetic changes, that all these mechanisms were already in place for the previous common ancestor who would not have had the proteins encoding for the hormone Leptin on the genome. So by that logic, the same processes should have happened for the mouse genomes, and to say that they wouldn’t, would be special pleading. If I were an evolutionist, before the genomes were sequenced, I would have predicted that the sequence difference between mammals like mice and humans, in the 3-d protein coding sections for the hormone leptin in the OB gene, should be proportional to the amount of time that they last had a common ancestor, because of mutational load over time. But according to research currently done, they are very similar. By this point, the portion of the genome of the mouse that codes for Leptin should have been changed by mutation into another protein coding for something else. So this makes alot of sense that certain portions of the genomes would be similar or identical, even from long distance creatures from the evolutionary time scale, because all animals need to perform certain functions, from bacteria to humans, to live, but only if this was a SINGLE designer putting his copyright on his design.
I read your article on your website, while I found most of what you said to be of good report to figure out what the results of a 70% overall similarity, according to Tomkins study, as well as proposing other research to see if the results support an evolutionary or creationary viewpoint, I am surprised that you are still saying that, “portions of the genes are huge tracks (hundreds of millions of base pairs) of the human genome that are repeated sections and pseudogenes (broken, unused genes) and those regions have very low sequence similarity to other animals.” This would beg the question of how you know that these sequence pairs are broken or functionless? These are the exact types of arguments that were used for organs in the body, they are “broken” organs, or “unused” organs. I find this view to be unscientific. We should wait to see if these genes are in fact the very reason that we are so different from each other in so many ways, as well as so different from similar looking creatures and not just assume that they are all functionless. We have all been able to see just how different two individual human males can be, or two individuals period, sometimes even between twins I have seen radical differences in personality, etc. It seems unwise to repeat the same mistakes of the past over again. I can’t agree with you that if we include repetitive DNA, and large changes in spacer sequence sizes like Tomkins that the similarity number will fall to the 80s at best. I think that these repetitive sequences that are currently assumed as pseudogenes, are actually functional and contribute to what a creature ends up being. Like I said before, only more research will be able to clarify this. Like I said before, I am not a geneticists, so the more information I can get to discern weather my current views are correct or not, the better, because I actually really only want to know what the best possible solution is at this point. I am so secure in my faith that the Creator God of the Bible is who made everything, that I am only after what is the best solution.
Hi Eric, good questions and points. I appreciate the dialogue and don’t’ want to wear out my welcome here but let me try to give as short of a response as I can. Regarding the sequences of something like leptin, genetic load doesn’t really matter. Mutations don’t build up in a gene like leptin because mutations that change an important position are lethal and so only members of a population that have the functional version of the gene will survive. You are right that to expect that mutations would accumulate from the time of split with a common ancestor. That was part of the idea with the mitochondrial genomes from the article you read. What I think might not be apparent is that mutation only accumulate at an even rate over time if there is no selection on that sequence. If there gene needs a particular sequence then selection will eliminate any individual with the mutation and keep the ones without the mutation so you won’t see the mutations accumulate over time at a very fast pace. On the other hand portions of DNA that aren’t used or have no selection (like many third base positions in genes) are free to vary and so will accumulate mutations but these don’t affect the protein shape. The leptin example isn’t that great because this region of the genome is so similar but I crunched the numbers from an alignment I have. The leptin gene is about 500 bp in length (same in almost every mammal) and human and chimp line up easily and I see only 2 differences (<0.5%) in the coding sequence (the part that codes for the amino acids of the hormone). Those two difference I think are synonymous (are at redundant codon positions) so if I remember right chimps and humans make exactly the same leptin hormone. These sequences are somewhat constrained from change so this is not surprising however as many as 15% of the DNA sequence could be altered without effecting the protein sequence at all so there isn't any a priori reason to believe that humans and chimps need the same DNA sequence. This gene also has a 2242 bp intron in the middle of the coding sequence. Both chimps and humans have this intron at the same position. I count 22 differences in this region which is a 1% difference. I also looked downstream of the gene so beyond where the gene code ends and there is “spacer” sequence and for the next 2000 bp I see about 30 differences which is a 1.5% difference and I also see a few small insertion and deletions of bp so maybe 3% total difference. The point is not the overall difference but that sequences that are in gaps between coding sequences would be predicted to have more differences because they are expected to accumulate mutations consistently over time. As a reference, the difference between a chimp and great ape is only 4 differences in the coding sequence and at least 35 differences in the intron. I see in the ARJ that Jean Lightner is calling chimps and apes the same kind and implies there was only one pair of great apes on the ark. If this is so all of these differences then resulted from mutations from the common ancestor. It is understandable that evolutionist would look at that argument and say, but the human and chimp have fewer differences. I have a hard time telling them their logic is faulty. Of course this is just in one gene and that doesn’t tell the whole story but the differences one sees in the same “kinds” aren’t what one would expect if they have a close genetic connection.
As an aside, leptin is found in all vertebrates except for birds where it has been found to be utterly lacking. Yet birds have a receptor for the hormone that works if you inject human leptin into the bird. It seems birds must use another protein of completely different DNA sequence to make the same shape protein (at least the shape where it should bind) to bind to the receptor. The space in the genome where the gene should reside (compared to the order of genes in other reptiles and mammals) is just missing the DNA code for leptin. There is a lot of effort being put into finding out how birds signal their leptin receptor in these birds. Wow, been going on toooo long again. Not sure I am very clear about this stuff but hopefully it is at least interesting. I've been having fund playing with the sequences. Joel
Hey Joel, thanks for the response. While I do find the talks of genetics to be some what confusing, you did a good job explaining things and I did find what you said to be interesting!
I find that very fascinating that birds do not have the Leptin sequence that most vertebrates have and yet they respond to human Leptin. I wonder just how different their genetic sequence that codes for the Leptin protein receptor(?) is to the other creatures and why they would differ from both a creationist as well as evolutionists perspective.
Hi Eric, the leptin bird story is an interesting study in how discoveries are made in science. It isn’t always straightforward. There was a Chinese group that reported a leptin sequence many years ago and then many studies followed that attempted to show how leptin could be used in chickens to alter fat content in production. However, other leptin researches noted that the leptin sequence that this group reported was barely different than mammals. The Chinese group really never gave any thought to the expected sequence of a group that was not a reptile or mammal and so just assumed they had the bird sequence. I suppose an old age evolutionist or a young age creationists might both expect that there would be large difference in the sequences. The former because of time of divergence from common ancestor and accumulated mutations, the latter because of lack of common design in birds vs mammals. Further attempts to find leptin in birds resulted in no further discoveries or duplications of the first report and now it is assumed that that chicken “leptin” sequence was a contaminant from the lab (a lab that worked with mammal leptin). Now that the entire chicken genome has been sequenced and other bird genomes they have been scoured for any sequences that had any likeness to leptin and none have been found. If it were there someone would have found it because there is intense interest in this gene by the poultry industry and so anyone who finds leptin would have a “nature” paper and could get rich off studying the gene. But you asked about the receptor. That is where it gets really fascinating. The leptin receptor in birds has very little sequence variation and thus is considered a “conserved” gene and the receptor sequence really is not much different than reptiles and mammals which also have very similar sequences. Protein models of the binding locations for the leptin hormone reveal virtually identical protein shapes and amino acid codes for the binding site. The only place the bird sequences differ end up resulting in changes in the peripheral parts of the receptor that doesn’t seem to have any function significance. This is really unexpected though fits with the observation that human and other mammalian leptin can trigger this receptor in birds.
So what does a special creationist and evolutionists with this receptor? The evolutionist assumes that since reptiles and “lower” animals have leptin that the ancestor of birds had leptin but some other gene also produced a protein of similar shape that under some conditions was able to bind to the leptin receptor in birds and also trigger the response (there are many effects of binding – lower apetite, bone density changes etc..). With another gene also able to trigger the response this formed a redundant system. Such redundancy meant that any mutation that caused the leptin not to function properly would not be lethal to the organism as it would be in you or I or any other mammal. Once the gene function was destroyed and thus became a pseudogene the common ancestor of birds had a mutation that eliminated it from the chromosome altogether. Currently the thinking is that this other gene in birds must produce a similar shaped protein that is playing the role of leptin so there is a search for this other gene predicted to exist going on right now. The creationists perspective would probably be that birds have to have a fat lipostat/controller just like mammals and so they have a receptor protein in their brains that reads the level of fat in their system but rather than the lepin gene He created a different gene with the same binding site as leptin but He kept the receptor the same as in other vertebrates. It could be the receptor also reacts to other hormones we don’t know about yet and so has some other reasons for being the same in all vertebrates. I think that even the creationist would predict, based on the presence of the receptor, that a leptin-like hormone should be present in birds and a search for this hormone will eventually reveal another gene that is acting like leptin in birds.
Dr Wile, just to clarify:
‘The only comparisons I see deal with genes. While this is interesting, it has little bearing on whole-genome comparisons, since genes make up such a tiny percentage of the entire genome. ‘
Indeed. But genes would be the regions of the genome we would expect to me the most conserved. Non-coding regions will have much lower similarity between populations than coding (‘gene’) regions.
So your argument here is actually backfiring: Exactly because we already see a difference in gene sequence between cow and yak that you said you would be ‘extremely surprised’ to see, we can expect that including non-gene regions would exacerbate this problem, not solve it.
‘They are quite different. Look, for example, at Supplementary Figure 10. In the case of the yak and cow (graphs A) the number of genes that are highly similar is significantly greater than the number of highly similar genes for the human and chimp (graphs B).’
How do you get the idea that it is ‘significantly greater’? The term ‘significantly greater’ would imply that there are some statistical analyses you performed – is that so? Are they included in the paper?
Moreover, the figure you reference simply gives the raw number of genes with similarity – it doesn’t give the proportion of genes with similarity. Your argument would only make sense here if the proportion was greater in yak/cattle, but the figure gives no indication of how many genes were actually compared to obtain the figure.
Finally, if you actually read the sentence in the article referencing that figure, the very next few sentences completely contradict your overall argument:
‘Average synonymous (Ks) and nonsynonymous (Ka) gene divergence values between yak and cattle were 0.0114 and 0.00207, respectively, close to the values between human and chimpanzee genes (Supplementary Fig. 11). Yak and cattle were estimated to have diverged approximately 4.9 million years ago, which is comparable to the time at which humans and chimpanzees diverged (Supplementary Fig. 12).’
Thus, the authors quite unambiguously state that the similarity between yak and cattle is very similar to that between chimpanzees and humans, which fits perfectly with an evolutionary model, but not with the baramin model which, as you rightly say, would expect much greater similarity between yak and cattle than between humans and chimps.
Singring, you couldn’t be more wrong in your assertions. First, there is no reason to expect that the protein-coding regions of DNA will be the most conserved. There is an enormous amount of conserved noncoding DNA out there as well (see here, here, and here, for example).
Second, I never said I would be surprised to see a difference between the genes of yaks and cattle. I said, “I would be incredibly surprised if the cow and yak genomes are only about 80% similar.” The paper specifically says, “Overall, yak and cattle genes were highly similar, with 45% of encoded proteins identical and mean protein similarity approximating 99.5%.” (emphasis mine) This is higher than the protein similarity between chimps and humans, so even when we restrict ourselves to just the genes, we see that the creationist expectation is fulfilled.
Third, you can easily do the statistical calculation yourself to see that the similarity between yaks and cattle is, indeed, significantly greater than that between humans and chimps. For example, look at the number of genes that code for the identical set of amino acids in supplementary figure 10 (0 amino acid difference). In cattle and yaks, it is roughly 2,700. In humans and chimps, it is 2,500. The statistical error for 2,500 is +/- the square root of 2,500, which is 50. Thus the similarity is greater in yaks and cattle by 4 times the statistical error. For the genes that code for only one amino acid that is different, the number of genes is higher in the yak and cattle by more than 15 times the statistical error. So yes, the yak and cattle are significantly more similar than the chimp and human. Also, the paper specifically says that the gene count in yaks is estimated to be 22,282. This is similar to the gene count in humans. Thus, you can compare the raw numbers, since the total number of genes is roughly the same.
Fourth, the quote you give does not contradict my argument at all. The quote simply gives the averages, and it only says that those averages are “close” to the values of chimps and humans (which are not given). All you have to do is actually look at the figure the quote references (Supplementary figure 11) to see how radically different the Ka, Ks, and omegas are in the two comparisons. Notice that for the yaks and cattle, the Ka peaks at about 0.0017, while for the chimp and human, it peaks at about 0.0024. More importantly, look at the difference in the shapes of the graphs. The yak/cattle graph peaks early with a long tail, while the chimp/human graph is nearly a bell curve. Similar differences can be seen in the Ks graphs. Now look at the omega graph. For the yak and cattle, it peaks at about 0.18, while for the chimp and human, it peaks at about 0.24. Since the range of the yak/cattle omega graph is from 0.1 to 0.3, this means the peaks of the two graphs are different by about 30% of the yak/cattle range. That indicates significant difference between the two comparisons.
As you can see, then, these data support the creationist expectation that the yak and cattle are genetically more similar than the chimp and human. Once again, of course, this isn’t all that illuminating, since only genes are being compared. Nevertheless, the analysis that does exist is (not surprisingly) exactly what a creationist would expect.
Dr. Wile,
Thank you for the very informative and thought-provoking article! I am very curious as to the method by which the DNA is analysed for these studies. With such extremely small material, how is this accurately “chopped” up in an organized manner? Also, how can the analyst tell which is the exactly corresponding bit of DNA from the other creature so as to make the comparison? Thank you for taking the time to answer comments on your blog!
Thanks for the questions, Enoch. The answers are pretty detailed, and I think this article by Dr. Tomkins provides them.
‘First, there is no reason to expect that the protein-coding regions of DNA will be the most conserved. There is an enormous amount of conserved noncoding DNA out there as well.’
If I ma quote from each of the three studies you reference to support your claim:
First paper: ‘In general, similarities in sequence between highly divergent organisms imply functional constraint.’
Second paper: ‘Large-scale conservation of non-coding genomic regions has been discovered by Dermitzakis et al, after alignment of the human chromosome 21 to homologous regions of the mouse genome. This work reported that protein-coding genes were more conserved overall than non-genic regions, thus giving a large-scale confirmation that evolutionary conservation is a hallmark of biological function.’
Third paper: ‘Using a comparative genomics approach, Dermitzakis and colleagues have recently shown that at least some non-coding sequence, frequently ignored as meaningless noise, might bear the signature of natural selection.’
All three papers state that these findings of conserved non-coding regions are exceptions and that the general picture is one of non-coding regions being less conserved that coding regions. In fact, much of molecular taxonomy is based on this difference.
‘This is higher than the protein similarity between chimps and humans, so even when we restrict ourselves to just the genes, we see that the creationist expectation is fulfilled.’
See my quote from the paper – the authors explicitly state the opposite of what you are asserting here (more on this below).
‘Third, you can easily do the statistical calculation yourself to see that the similarity between yaks and cattle is, indeed, significantly greater than that between humans and chimps’
I’m afraid you seem to have a completely misguided idea of how statistical analysis works. Your statistical error is completely inappropriate for this kind of comparison for several reasons. First, the data we are dealing with here is categorical (categories of gene similarity). Calculating a statistical error is not appropriate for this kind of data – it literally is like trying to measure the weight of something using a ruler – two completely separate things. Errors are calculated for continuous data where a mean can be calculated. This is not the case here.
Second, even if you could use a statistical error here, you would need to know the number of samples involved – as I have already pointed out to you, maybe you missed it.
Imagine I presnted you with data saying that I found 100 women with an IQ of > 120, but only 16 men with an IQ or > 120. Using your ‘method’, you could see that the difference in these data is almost eight times the ‘statistical error’ for women, which would mean that women are significantly more intelligent than men, right?
Well, what if I had taken data for 200 women and 32 men? Then the proportions of individuals with an IQ > 120 would be exactly teh same for both – 50 %.
So on the face of it – your ‘statistical analysis’ is inappropriate for these data. But what are the actual data. Well, if you look at supplementary figure 8 for the paper in question, you will see that 8923 genes were compared for yak and cattle, but only 7499 were compared for humans and chimps. Going back to supplementary figure 10, we can generously estimate that about 2800 genes were in the category ‘very similar’ for yak and cattle, but only 2500 genes are in that category for humans and chimps. Something you called a ‘significant difference’.
The appropriate test to use on categorical data like these is a Chi-Square test, which takes into account the total number of genes being compared. So, I ran a Chi-Square test on these data, and the result was this: The preportion of genes with ‘high similarity’ is actually *higher* for chimps and humans (33.33 %) than it is for Yaks and cows (31.37 %). The Chi-square value was 7.148 for a P-value of 0.008. A P-value of less than 0.05 indicates a significant difference between data.
So: not only is the trend the opposite of what the baramin model would predict it to be, this difference is actually significant in exactly the opposite way you claimed it would be.
‘All you have to do is actually look at the figure the quote references (Supplementary figure 11) to see how radically different the Ka, Ks, and omegas are in the two comparisons…That indicates significant difference between the two comparisons.’
Once again your use of the term ‘significant difference’ id completely inappropriate here. Seeing as how flawed your statistical analysis of these data is, I think you will understand that I take the rigorously peer-reviewed analysis of several authors in Nature over your assertions as to what you see when you ‘look’ at the graphs.
Singring, you seem to be confused on several levels. I hope I can clear up some of that confusion for you. The point of the papers I linked to you was to indicate that until you actually study the noncoding DNA, there is simply no way to predict how much it is conserved. Remember, the title of the third paper is “Unexpected conserved non-coding DNA blocks in mammals.” Since the noncoding regions in yaks and cattle have not been compared, there is simply no way to determine whether it will be more or less similar than the genes. I agree that the evolutionary expectation is that they should be less conserved. However, the evolutionary expectation was that noncoding DNA should not be conserved over a wide range of species.
You claim that the authors explicitly state the opposite of what I am saying. However, they explicitly state the opposite of what you are saying. Once again, the authors specifically state, “Overall, yak and cattle genes were highly similar, with 45% of encoded proteins identical and mean protein similarity approximating 99.5%.” However, only 29% of proteins are identical between humans and chimps, and even evolutionists say that the similarity between protein-coding genes in chimps and humans is only 98-99%. Thus, the authors explicitly state that the genes in yaks and cattle are more similar than the genes in humans and chimps.
Your quote says nothing about relative similarity. It only discusses the averages, which are called “close.” Please review what I tried to explain to you previously, because if you look at the actual graphs upon which that quote is based, you can easily see how radically different the comparison between yaks and cattle is compared the the comparison between humans and chimps. Perhaps you missed that explanation.
I did not miss your attempt to “point out” the number of samples involved. As I explained to you, the gene count in yaks is about the same as that found in humans, so we know that the sample sizes are roughly the same. Perhaps you missed that. This, of course, is why your example of the IQs of men and women doesn’t apply to this discussion at all. Since we know the gene sample size is roughly the same, we know that it is fine to compare raw numbers. This, of course, is why the statistical analysis I used is also totally applicable.
Now…let me clear up your confusion about Supplementary figure 8. You seem to think that the figure indicates the yak/cattle comparison was made based on 8,923 genes, while the chimp/human comparison was made based on 7,499 genes. That’s not what the figure says at all. The figure says that they found 8,923 genes in the yak genome that are orthologous to gene in BOTH the cattle and human genomes. It then says that 7,499 of those genes are also found in the chimp. There is no indication that these are the genes compared in Supplementary figure 10. If so, then the comparison tells us nothing, because it is only concentrating on orthologous genes found in all four species. It tells us nothing about how similar the human and chimp genes are overall. If this is the case, then the Chi-Square analysis you made is not applicable for indicating the relative similarities, as it is not comparing all orthologous genes in the chimp and human genomes.
In fact, we know for certain that your Chi-square analysis is wrong, because as I quote above, we know that 45% of the genes code for identical proteins in yaks and cattle, while only 29% of genes code for identical proteins in chimps and humans. This is precisely opposite of what you are claiming with your Chi-square analysis.
When I looked at the study, I assumed that Supplementary figure 10 showed how all orthologous gene varied. If my assumption of what genes are used is correct, then as I mentioned to Joel, the graphs show significantly more orthologous genes between yaks and cattle than there are between chimps and humans. Since the total gene count is roughly the same among all the species, this tells you once again that yaks and cattle are significantly more similar than apes and humans.
No matter how you slice it, then, the data conform to the creationist expectation: even if you restrict the discussion to genes, yaks and cattle are significantly more similar than humans and chimps. I do hope I have cleared up some of your confusion on this matter.
Hi Dr. Wile, I think that if you are expecting that the differences will not be that great then you are going to be left disappointed in the future. This was mostly why I originally commented. I’m afraid that Christians are putting too much hope in obvious genetic differences as proof of special creation. Finding that evidence in DNA is going to be a difficult sell. From what I see Tomkins and other are solely trying to sell it to the internal Christian audience but if a Christian finds comfort in these numbers and then goes on to study genetics they are going to find themselves ill-prepared to deal with the data that they find. I feel very confident that using Tomkins methods will result in vast differences being found between most species on earth and once thousands of comparisons are done, the differences between humans and chimps will looks much less significant than that 70% would make them appear today. That would be very unfortunate in my mind as overall similarity or difference is a poor method of comparison. Its all about very small but very important differences that can make species very different from one another despite very similar genome compositions. What do those few individual differences in the protein coding genes do rather than those big differences in the non-coding sequence that likely have little meaning to the morphology of the organism. The focus should not be based on these rough similarity matrices. That said, the Yak/cow comparison can be done despite what Tomkins says, I understand that there are portion of the genome that have not been constructed to high confidence but there is a large enough portion of the genomes that can be compared that the same test could be applied. In the end Tomkins could come up with a number that represents the minimum similarity but because some large portions of the YAK and cow sequence can’t be aligned, obviously these regions have much lower similarity and so the actual overall similarity would be less than the number he derives but this would still be a useful comparison. I have played with some genome aligners and the YAK and Cow genomes are significantly different from one another in non-coding non-intron space. You are right that some non-coding regions are highly conserved but those are very special cases and do not represent the typical non-coding space especially the non-intronic portions. You might also be confounding protein coding similarity and DNA sequence similarity. The 99.5% is protein coding similarity which is less than 0.1% difference from chimp/human protein similarity. Human/chimp and cow/yak coding DNA sequence similarity are both lower than that number. All that said, I probably wouldn’t focus on cow/yak but rather on fox/dog, tiger/lion or sheep/goat all of which I think are more different than yak/cow but for creationists are each one kind.
BTW, I will be attending at least the first day of the International Creation Conference in August. One talk that I will attend will be Todd Woods about mitochondrial genome comparisons in canines and felids and what it means for baraminology. That should be very interesting because I do trust Wood to present the raw numbers accurately and he isn’t afraid to say when something isn’t what creationists would expect. I might not agree with his interpretation of how we might then explain the data but at least I feel like he gives the data a chance to be heard and it leads him to actually deal with it. For Tomkins I feel like he wants to find differences and has found a method to suit his purposes but he has to be willing then to let his method lead him to look at other data and deal with whatever it may say. Wood should find the same differences I did in my comparison of whole mtDNA alignments. It will be interesting to see what he says about foxes and dogs and cheetahs and domestic cats having more differences than chimps and humans. I think he may conclude that foxes really are a different baramin although I can think of a couple other possible explanations for the differences.
I appreciate your concern, Joel, but if I am disappointed, I can handle it. After all, my real goal is to understand what the data are telling us. I am excited to find the real answers. This is why I specifically asked Dr. Tomkins to do a similar analysis on animals that are thought to be a part of the same monobaramin. I think it will be an excellent test.
No, Christians who take these numbers seriously are not going to be “ill-prepared” for what they find when they study genetics seriously. In fact, those who believe what we find in the popular media (that humans and chimps are 95-99% similar genetically) will be unprepared for what they find when they study genetics. As a review article in the Journal Science demonstrates, when topics are learned in the format of a controversy, they are learned better. In the end, Christians who learn about studies like those of Tomkins (and others) will probably be better prepared when they investigate genetics in more detail.
I understand that you feel very confident that when a “Tomkins analysis” is done on a lot of organisms, the difference seen between chimps and humans will not look that great. However, I feel very confident that this will not happen. Right now, we simply do not have the data to determine which feeling is correct. However, based on the yak study you posted, the protein-coding genes conform to the creationist expectation. I understand that this is only part of the genome. In fact, I pointed that out as soon as you posted the study. However, it’s all the data we have right now, and those data show that yaks and cattle are significantly more similar than humans and chimps when it comes to the protein-coding genes.
Where do you get the number of 0.1% difference between human and chimp protein-coding genes? That number makes no sense to me at all. Did I misunderstand what you wrote? After all, we know that 45% of proteins are identical between yaks and cattle, while only 29% are identical between chimps and humans. In addition, every analysis I have seen indicates that the protein-coding genes are about 98% similar between chimps and humans, not 99.9% similar.
I agree that genomes like the fox/dog or lion/tiger would be more interesting. Right now, however, those genomes have not been sequenced. Since I thought your suggestion of comparing animals within a monobaramin with a “Tomkins analysis” was interesting, I wanted it to be done soon, and the yak and cattle genomes have already been sequenced.
I would strongly disagree with your comparison of Dr. Woods and Dr. Tomkins. I think they are both trying to be as honest as they can be with the data. However, as I stated previously, since we don’t really understand genetics very well, it is difficult to know how to properly compare genomes. Dr. Woods and Dr. Tomkins simply are doing it in different ways. The more we have such different analyses, the more likely we will be to figure out the proper way to make such comparisons.
‘However, the evolutionary expectation was that noncoding DNA should not be conserved over a wide range of species.’
Dr Wile, I urge you to be accurate in your use of language: The evolutionary prediction is that non-coding DNA is less conserved than coding DNA – not that it is not conserved at all.
‘The point of the papers I linked to you was to indicate that until you actually study the noncoding DNA, there is simply no way to predict how much it is conserved.’
The papers unambiguously state that non-coding DNA is generally less conserved than coding DNA, exactly in line with evolutionary predictions. I really don’t see how anyone could state it any clearer than in paper 2:
‘This work reported that protein-coding genes were more conserved overall than non-genic regions, thus giving a large-scale confirmation that evolutionary conservation is a hallmark of biological function.’
Next, you go on to compare apples with oranges:
‘However, only 29% of proteins are identical between humans and chimps,… ‘
The study you link to is a genome-wide study for humans and chimpanzees. The figures we are discussing here from the yak paper do NOT cover the entire genome. The authors explicitly state this in the methods, twice:
‘First, we used TreeFam20 to identify 13,810 homologous gene families shared by 4 species (yak, cattle, human and dog)…’
‘We used conserved genome synteny methodology to establish a high-confidence orthologous gene set that included yak, cattle (UMD 3.1), horse (EquCab2.0), dog (CanFam2.0), mouse (mm9), chimpanzee (panTro2) and human (hg19) genes.’
They worked on subsets of orthologous genes when conducting their comparisons, not the full genome.
This should be obvious if you look at Figure 1, which gives a figure of about 16,000 genes for yaks. However, if you tally up the number of genes in supplementary figure 10, it falls way short of that number (at about 7,800 genes). So obviously that figure is not referring to a genome-wide comparison of genes, but only to a shared subset the authors investigated! This makes sense because to compare between different groups, it makes no sense to use genes that are present in one but not the other.
‘The figure says that they found 8,923 genes in the yak genome that are orthologous to gene in BOTH the cattle and human genomes. It then says that 7,499 of those genes are also found in the chimp.’
EXACTLY. But it is these subets of genes the authors based their further analysis on (at least this is what I am forced to assume, the methods are incredibly condensed). What supplementary figure 10 is clearly *not* based upon is a full-genome compariosn, because the numbers don’t even match up with Figure 1 in the paper proper.
‘If this is the case, then the Chi-Square analysis you made is not applicable for indicating the relative similarities, as it is not comparing all orthologous genes in the chimp and human genomes.’
1.) Once again, figure 10 is clearly not referencing a full-genome comparison.
2.) If we are in disagreement over the source data for figure 10, there is a simple way to settle this: just tally up the number of genes actuall presented in figure 10. If you do that, you will come up with (approximately) 7,750 genes in the yak/cow comparison and (approximately) 5,000 genes in the human/chimp comparison.
Once again, we can do a Chi-Square test just on this data, comparing the proportion of this total number of genes that was compared for each pair that were identical (no difference in amino acide sequence). Once again, that would be 2,800 for yak/cow and 2,500 for humans/chimps (approximately).
The result: A whopping Chi-square of 240 and a P value less than 0.001, with yak and cow showing about 36 % identical genes, versus 50 % similarity for humans and chimps!
You can see this pattern clearly reflected in the graph – the slope for decreasing similarity is much stronger in the human/chimp graph compared to the yak/cow graph. This pattern would hold up no matter what ‘total number of genes’ we scale it up or down to and the results of the test would still apply.
So this again completely goes against your assertions.
Singring, I agree that evolutionists are now admitting that noncoding DNA can be conserved across many species. However, that was not the original expectation. Indeed, as I pointed out before, one of the papers I linked was entitled, “Unexpected conserved non-coding DNA blocks in mammals.” (emphasis mine). The point is that since evolutionary expectations have been shown to be incorrect in the past, it would not be surprising to see more evolutionary expectations shown to be incorrect in the future. Thus, I will wait until a the noncoding DNA in yaks and cattle are compared before I decide what that comparison will show. It seems to me that’s the more scientific attitude to take.
I think you are correct about the comparison that was made. The authors are only looking at the genes shared in all four species. Since that is the case, it tells us nothing about the relative similarity of yaks and cattle as compared to humans and chimps. After all, if we are asking about how similar the genes in chimps and humans are, we must look at all the genes. Since the authors are not doing that, it tells us nothing about how much more or less similar yaks and cattle are compared to humans and chimps.
So yes, I was comparing apples and oranges. However, so are you. You are claiming that this paper shows yaks and cattle are less similar in terms of their genes than are chimps and humans. That is certainly not true. All it is saying is that if you consider the small fraction of genes that are orthologous in all four species, the differences between the two are close. That doesn’t relate to the creationist expectation at all.
The creationist expectation is that the entire human/chimp genomes are significantly less similar than the entire yak/cattle genome. Since no one has done a full comparison of the genomes, we can at least look at the genes to some idea of the similarities. If we look at the genes, we find that genome-wide, 45% of yak and cattle genes are identical, while only 29% are identical in chimps and humans. In addition, we find that genome-wide, the yak and cattle genes are 99.5% similar, while genome-wide, human and chimp genes are only about 98% similar. Either way you look at it then, based on genes, humans and chimps are less similar than yaks and cattle, exactly as the creationist would predict. I suspect that if a full genome comparison is done, it will also confirm the creationist expectation.
I do appreciate you pointing out that the paper’s results really don’t apply to this question at all. I had clearly misread Supplementary graph 10, and I am glad that you corrected me on that.
Dr. Wile, Thanks for your willingness to work with comments on your blog. I know how much effort that takes. This has been helpful to me. I think that we will learn much in the next few years and will have the kinds of meaningful comparisons that we need to make soon. Regarding the 0.1%, I wasn’t clear. I intended that the 0.1% different was different from the 99.5% (so 99.6% for human/chimp) similarity from Yak/Cow shared proteins sequences. These numbers get confusing because there is not only protein similarity vs DNA similarity but orthologous gene similarity vs similarity in gene families or similarity of non-coding vs coding etc.. I get myself confused trying to remember all the different estimates and how they were produced.
Thank you, Joel, for being so pleasant and engaging. I know there are many things you and I disagree on, and it is so nice that you interact in a very pleasant way, regardless of our differences. I do agree that as time goes on, the comparisons we make between organisms will become more meaningful.
I still don’t think I understand your number when it comes to gene similarity. I don’t know any recent data that indicate humans and chimps are 99.6% similar in protein coding genes or the proteins themselves. Even Dr. Moran says the gene similarity is only 98-99%. If you look instead at the proteins themselves, there seems to be even more difference than what you see in the genes. One study says that 80% of proteins differ between chimps and humans. Another study indicates there are 689 genes in the human genome not present in the chimp genome and 729 genes in the chimp genome not present in the human genome. Assuming each gene represents only one protein (which we know is wrong because of alternative splicing), this indicate a protein similarity of only 94% (1-[689+729]/23,000)x100.