Before the human genome was sequenced, it was thought that humans had well over 100,000 genes. This reasonable conclusion was based on the fact that that the human body is estimated to produce 120,000 – 140,000 different proteins. Since biology had determined that a gene tells a cell how to make a protein, it was assumed that 120,000 – 140,000 proteins would require 120,000 – 140,000 different genes.
As is often the case with science, however, the data turned out to be very surprising. When the human genome was initially sequenced, it was estimated to contain about 30,000 genes. Today, it is thought that the human genome contains 20,000–25,000 genes.1
So if a human cell requires a gene in order to make a protein, and if the human body produces as many as 140,000 different proteins, how can it do so with “only” 20,000–25,000 genes? A large part of the answer to that question has to do with an amazing process called alternative splicing.
To understand alternative splicing, you need to first understand what a gene is and how it instructs the cell to make a protein. A gene is a stretch of DNA that essentially has a recipe for making a specific protein. The stretch of DNA is copied (actually a negative image is made) by a similar molecule called messenger RNA (abbreviated as mRNA). That mRNA then leaves the nucleus (where most of the cell’s DNA is found) and takes the copied recipe to a ribosome, where the protein is made according to the recipe found on the mRNA. That’s the overview of how a gene tells a cell to make a specific protein.
There are a few things I glossed over in that quick explanation, but I want to highlight one of them, because that’s where alternative splicing comes in. It turns out that the genes in human (and all eukaryotic) DNA are made up of sections called introns and exons. Initially, RNA makes a copy of all sections. At that point, it is called “pre-mRNA.” Before it leaves the nucleus to go to a ribosome, however, the introns are removed, as shown in the figure below:
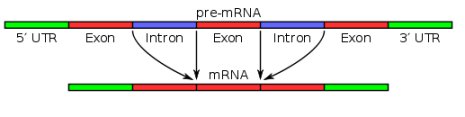
The blue sections in the illustration represent introns, and the other sections represent exons. Before the pre-mRNA becomes mRNA and can leave the nucleus, the introns (blue sections) are removed, so that in the end, only the exons are left. This, of course, led many researchers to think that introns were useless “junk DNA.” After all, what possible purpose could they serve if their copies were removed before the mRNA left the nucleus? This, of course, fit nicely with Dr. Susumu Ohno’s concept that sections of junk DNA were leftover “fossils” of extinct DNA produced by the evolutionary process. As a result, the idea that introns are junk became very popular.
We now know that this idea is simply wrong. At least one of the functions that introns serve is to separate the exons so that one gene can code for many different proteins. Remember, there are only 20 – 25 thousand genes in the human genome, but they code for 120 – 140 thousand proteins. This happens because the exons in a single gene can be spliced together in different ways. Consider the figure below.
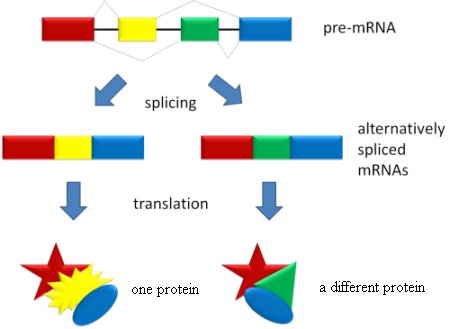
In the figure, the colored regions represent exons, and the black bars on the pre-mRNA represent introns. When the gene is originally copied, the introns and exons are both copied together, making the pre-mRNA. When the introns are removed from the pre-mRNA, the cell can use the exons as building blocks to make different proteins. In the illustration, the red exon, the yellow exon, and the blue exon are put together, making one protein. When the red exon, the green exon, and the blue exon are put together, however, a completely different protein is made. Thus, a single gene can produce more than one protein, because the exons can be put together in different ways. That’s alternative splicing, and it’s the major reason the human genome can have so few genes but code for so many proteins.
This, of course, is an amazingly elegant design. It takes a huge amount of information and compresses it into a small package. However, one important question should come to mind: “How often is this elegant design actually used?” In other words, do most genes participate in alternative splicing, or do just a few? The answer is we aren’t sure yet, but most studies indicate that up to 95% of the genes in the human genome participate in alternative splicing!2,3
Is that the only thing introns do? Are they just “spacers” that separate the exons so the exons can be spliced together in different ways? Probably not. You certainly wouldn’t expect that in such an elegantly-designed system. In the end, there are probably other jobs introns perform as well. As an article in the journal Science says:
Introns may play other vital roles, however. For instance, a slew of so-called small nucleolar RNAs (snoRNAs) are encoded by introns. Because snoRNAs accumulate in the nucleolus, where the protein-making ribosomes are formed, researchers speculate that they play a role in ribosome assembly.4
In addition, we know that certain introns play a vital role in the regulation of genes.5,6
So far from being the “junk” that many first imagined, introns are clearly a very important part of the genome. Indeed, as Dr. John S. Mattick, director of the Institute for Molecular Bioscience at the University of Queensland said:
The failure to recognize the full implications of this—particularly the possibility that the [introns] may be transmitting parallel information in the form of RNA molecules—may well go down as one of the biggest mistakes in the history of molecular biology
The more we learn about the genome, the less junk there appears to be. This, of course, supports the creationist view of the genome.
REFERENCES
1. International Human Genome Sequencing Consortium, “Finishing the euchromatic sequence of the human genome,” Nature 431:931-945, 2004 available online
Return to Text
2. Eric T. Wang, et al., “Alternative Isoform Regulation in Human Tissue Transcriptomes,” Nature 456:470-476, 2009 available online with subscription
Return to Text
3. Qun Pan, et al., “Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing,” Nature Genetics 40:1413-1415, 2008 available online with subscription
Return to Text
4. R Nowak, “Mining treasures from ‘junk DNA’,” Science 263:608-610, 1994 available online with subscription
Return to Text
5. L Ottavio, et al., “Importance of introns in the growth regulation of mRNA levels of the proliferating cell nuclear antigen gene,” Mol Cell Biol 10:303-309, 1990
Return to Text
6. A. B. Rose, “Intron-Mediated Regulation of Gene Expression,” Current Topics in Microbiology and Immunology 326:277-290, 2008
Return to Text

Of course it does! And of course your post topic is one you’ve found on another creationist source: Matheson’s Intron Fairy Tale and follow-up by Richard Sternberg.
Are these on-topic enough for you: PZ, Larry Moran and the same Matheson of Sternberg’s tall tale.
You are right, Shooter. It does support the creationist view, and I did get the idea for the topic from reading evolutionnews.
Of course, you forgot to post the one that shows PZ, Moran, and Matheson are all wrong. I will gladly include it so my readers can see how poorly your sources do when it comes to the science in question.
I love it!
And you forgot to link to Larry Moran’s post that shows Jonathan Wells is wrong. But I won’t blame you for that. It is not the responsibility of one side to quote the other. I, for one, am not going to wade through Evolution News and Views for relevant posts.
Shooter, of course you won’t read what is on evolutionnews, because you can’t stand to consider data that contradict your fervent faith.
Of course the link you posted is not a refutation of Wells. Since you are too afraid to read Wells, it is not surprising that you didn’t notice. All Moran did (as is typical) is try to distract from the issue at hand. He claims that Wells is too willing to believe the scientific literature when it is convenient for him. Of course, Moran is doing EXACTLY THE SAME THING. He chooses to NOT believe the studies that the vast majority of genes are involved in alternative splicing, because they contradict what HE wants to believe. He is free to do so, of course, but he shows his lack of logic by then complaining when others do the same thing.
So…we have a post from Wells that gives detailed references demonstrating that Moran is wrong, and you claim Moran “refutes” that by simply saying Wells shouldn’t believe everything he reads in the scientific literature. Wow! You really ought to read what you link!
I did read it. You need to read the comments section:
“The frequency of alternative splicing is a very controversial topic. It’s not possible to cite one or two references that settle the issue. One camp says that the vast majority of human genes are alternatively spliced and the other camp says that most of the data is artifact. According to them, the real percentage of alternatively spliced genes is much lower.
I really don’t care which side you support or which side Sternberg and Wells support. I think it’s an interesting and exciting debate.
However, I do care about whether you are aware of the controversy or are completely ignorant of its very existence. When you cite one or two papers supporting one of the positions then go on to base important calculations on those citations, then that indicates to me that you don’t understand the field.
Sternberg could have said something like this …
“There are several papers in the scientific literature claiming that 90-95% of human genes are alternatively spliced. While this data has been challenged, let’s assume, for the sake of argument, that it’s correct and do some calculations about the function of introns based on this maximum number.”
That would have been the correct thing to do if you know the field. Don’t you agree?”
Sorry about the forgotten end tag. Most of Wells’ argument is the niceness problem that we see so often leveled against the new atheists:
“But Matheson didn’t stop there. He demeaned Sternberg by calling him “poor Richard.” He also claimed that Sternberg is “disastrously clueless” because he doesn’t understand “the important and very basic distinction between a transcript and an intron.” Since every undergraduate biology student learns that an intron is a segment of DNA, while a transcript is a segment of RNA encoded by DNA, this last jibe is on a par with Moran’s insult that Sternberg can’t do elementary arithmetic. And it is equally unjustified.”
But poor Richard (I don’t know about you, but if someone compared me to Ben Franklin’s alter ego, I’d take it as a compliment) doesn’t know the very basic distinction between a transcript and an intron. And he made a very obtuse assumption – that every intron in a gene must be alternatively spliced – Moran didn’t say he can’t do elementary arithmetic. So what is an honest scientist to do with these mistakes, ignore them?
Remember, this whole “debate” is about an estimate. Sternberg says:
“This is just a rough estimate, of course. And as I wrote in my original critique of Matheson, even if I’m off by a factor of two we are still left with far more functional introns than Matheson acknowledges.”
Great, Sternberg has produced a larger estimate than Matheson. As Moran stated, lots of people have different estimates. Now what are we supposed to do about it? Moran, Matheson and Hunt are all mocking Sternberg for his understanding (or lack thereof, more precisely).
Shooter, you clearly didn’t read it until I challenged you, because you claimed that it refutes Wells. It doesn’t, not even in the comments section. The best that Moran can do is CLAIM (in the COMMENTS section) that the frequency of alternative splicing is “controversial,” but he gives no references to indicate it is. Remember, he wanted a number of 5%. He cannot cite a reference for that 5% number. Indeed, he cannot cite a reference for ANY numbers other than what Wells cites.
I think this comment from the comments section hits it on the head:
Indeed. What does Moran do in response to that comment? Does he give references? Of course not!
So…we have Wells’s article with a plethora of citations that back up every point made, and we have Moran’s article that just says, “believe me because I am an expert.” I know that kind of argument from authority is pleasing to you, but those of us who are actually interested in science know how empty it really is. Well done, Shooter. You have once again shown your reliance on the pronouncement of high priests instead of the evidence!