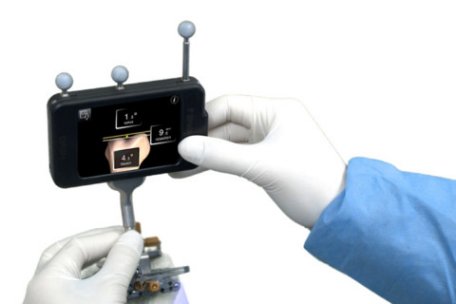
Last month, I wrote an article about an experiment involving spiders and an iPod touch. The results of the experiment were interesting, but I also thought it was really cool that an iPod was integral to the experiment’s design. Well, I just learned about something that I think is is even cooler. It turns out that an iPod touch is being used to assist in knee-replacement surgeries!
The system is called “Dash,” and it is made by a company called Brainlab. It consists of several accessories, such as probes, that attach to an iPod touch. Once everything is sterilized, the surgeon can use the probes to make measurements on the patient while the surgery is in progress. The iPod can then do some calculations on those measurements and show an image that will help the doctor install the artificial knee as accurately as possible. It can also use its WiFi capabilities to send those results to any other device, such as an iPad, if the surgeon wants a larger, more detailed image.
Why would a surgeon use a device like this? Well, in order for the replacement knee to function well, it must be aligned properly. In conventional knee replacement surgeries, the surgeon inserts a metal rod into the femur (the bone above the knee) to help with this alignment. Unfortunately, that process can increase the risk of certain side effects, such as fat embolisms. When using this iPod-based device, there is no need for an alignment rod. In addition, a surgeon who has been using it for more than a year says that the device provides better alignment than the conventional method. This leads to a larger range of motion for the artificial joint. Also, patients experience less pain and swelling after surgery.