As I posted previously, a huge leap in our understanding of human genetics recently occurred due to the massive results of project ENCODE. In short, the data produced by this project show that at least 80.4% of the human genome (almost certainly more) has at least one biochemical function. As the journal Science declared:1
This week, 30 research papers, including six in Nature and additional papers published by Science, sound the death knell for the idea that our DNA is mostly littered with useless bases.
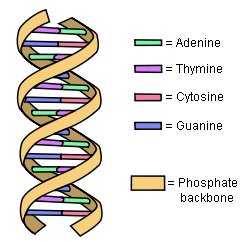
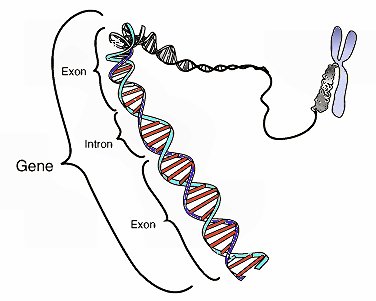
Not only have the results of ENCODE destroyed the idea that the human genome is mostly junk, it has prompted some to suggest that we must now rethink the definition of the term “gene.” Why? Let’s start with the current definition. Right now, a gene is defined as a section of DNA that tells the cell how to make a specific protein. In plants, animals, and people, genes are composed of exons and introns. In order for the cell to use the gene, it is copied by a molecule called RNA, and that copy is called the RNA transcript. Before the protein is made, the RNA transcript is edited so that the copies of the introns are removed. As a result, when it comes to making a protein, the cell uses only the exons in the gene.
By today’s definition, genes make up only about 3% of the human genome. The problem is that the ENCODE project has shown that a minimum of 74.7% of the human genome produces RNA transcripts!2 Now the process of making an RNA transcript, called “transcription,” takes a lot of energy and requires a lot of cellular resources. It is absurd to think that the cell would invest energy and resources to read sections of DNA that don’t have a function.
In addition, the data in reference (2) demonstrate that many RNA transcripts go to specific regions in the cell, indicating that they are performing a specific function. Since there is so much DNA that does not fit the definition of “gene” but seems to be performing functions in the cell, scientists probably need to redefine what a gene is. Alternatively, scientists could come up with another term that applies to the sections of DNA which make an RNA transcript but don’t end up producing a protein.
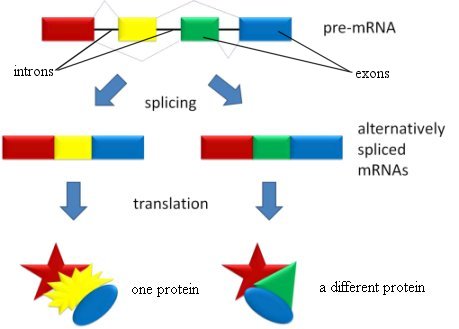
There is another reason that prompts some to reconsider the concept of a gene: alternative splicing. The ENCODE data show that this is significantly more important than most scientists ever imagined.