In a comment on a previous post about COVID-19, John D. said that he was watching Sweden and Denmark to evaluate whether or not shutting down most of society is an effective strategy against the disease. Why? Because they are very similar countries in the same basic region of the world, but they have remarkably different responses to the disease. Denmark has instituted many social-distancing strategies against the disease, while Sweden has not. Comparing how the disease is affecting those two countries might tell us something about how effective these strategies really are.
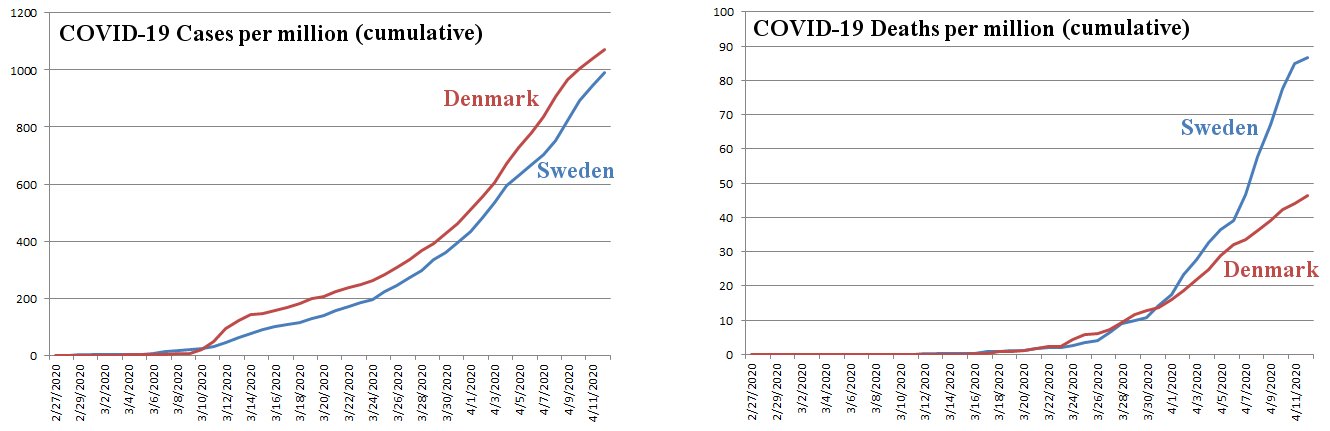
Well, I had a chance to look into this a bit, and the results of my analysis are shown in the graphs above. I got my data from the European Centre for Disease Prevention and Control. Of course, you could question the reliability of the source. However, I think that if it is not reliable, it is probably equally unreliable for both countries, so most likely, the comparison is justified. The data are compiled as a list of cases and deaths each day. I made a running day-by-day total of each and then divided by the population of each country in millions. So what you see in the graphs above are the cumulative number of cases (left) and deaths (right) each day, per million people in the country.
Looking at the graph on the left, we see that the rate of growth of cases is similar for both countries, but Sweden actually has a lower number of cases per million! So despite its social distancing guidelines, Denmark has more cases per million people than Sweden. However, look at the deaths per million, shown in the graph on the right. Not only does Sweden have significantly more deaths per million, they are increasing a lot faster than those in Denmark!
How can we understand the fact that Denmark has more cases but fewer deaths per million people? I personally think it’s because Denmark is probably testing more. Because of social distancing, doctors and hospitals are not doing a lot of the routine care they normally do. As a result, they are probably more focused on COVID-19, which probably results in more testing. It’s very possible that Sweden has a lot more cases, but since they aren’t testing as much as Denmark, that doesn’t show up. This is all just spectulation, of course. I don’t have the data to confirm whether or not Denmark is doing more testing than Sweden.
It’s also possible that less routine medical care in Denmark simply means that the people with COVID-19 are getting more medical attention, which leads to a higher rate of survival. So perhaps Sweden does have fewer cases, but since each case doesn’t get as much attention, the death rate is higher. Finally, it’s possible that because of social distancing, the sheer number of viruses to which each person is exposed is lower in Denmark. If that’s the case, the initial viral load on a patient is lower, which makes the disease more survivable.
Whatever the explanation for the fact that Denmark has more confirmed cases but fewer deaths per million, it appears that social distancing significantly reduces the number of deaths per million people in the population. Of course, I don’t think you can say that definitively based on this analysis alone, but the data do support that conclusion.
ADDED NOTE: If you look at the links in Dawn’s comment and Laree’s comment, you will see that Denmark is, indeed, doing more testing, which explains why they have more cases.